
This advance is independent of CDK activity regulation. This indicates that in the absence of these Ppases, critical CDK substrates can be phosphorylated at lower CDK activity levels (ie earlier), thus advancing mitotic entry.
This advance is independent of CDK activity regulation. This indicates that in the absence of these Ppases, critical CDK substrates can be phosphorylated at lower CDK activity levels (ie earlier), thus advancing mitotic entry.
Using a fluorescent sensor, we showed that without PP2A-B55, a candidate substrate (Cut3-T19) is phosphorylated earlier and less rapidly.
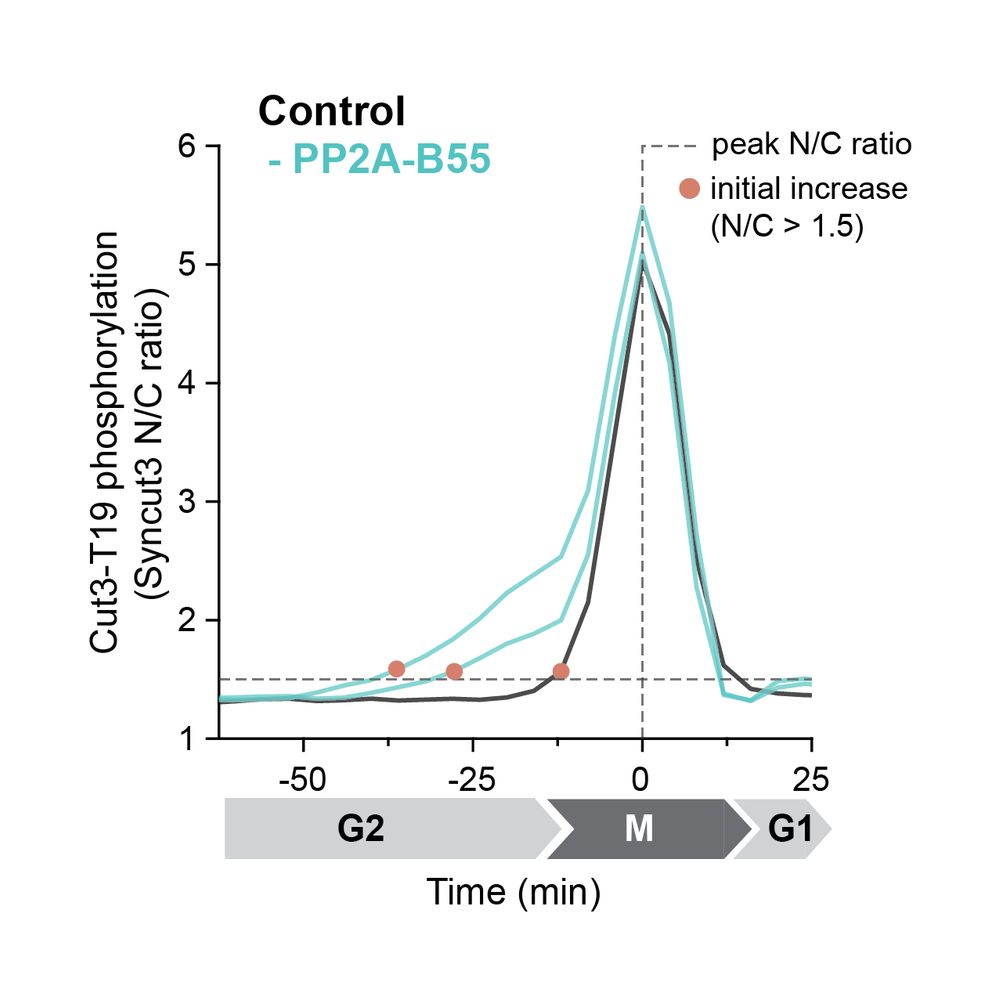
Using a fluorescent sensor, we showed that without PP2A-B55, a candidate substrate (Cut3-T19) is phosphorylated earlier and less rapidly.
Phosphoproteomic timecourse showed that CDK substrates targeted by CDC14 and PP2A-B56 are, on average, net phosphorylated first, followed by CDK substrates targeted by PP1 and then PP2A-B55.
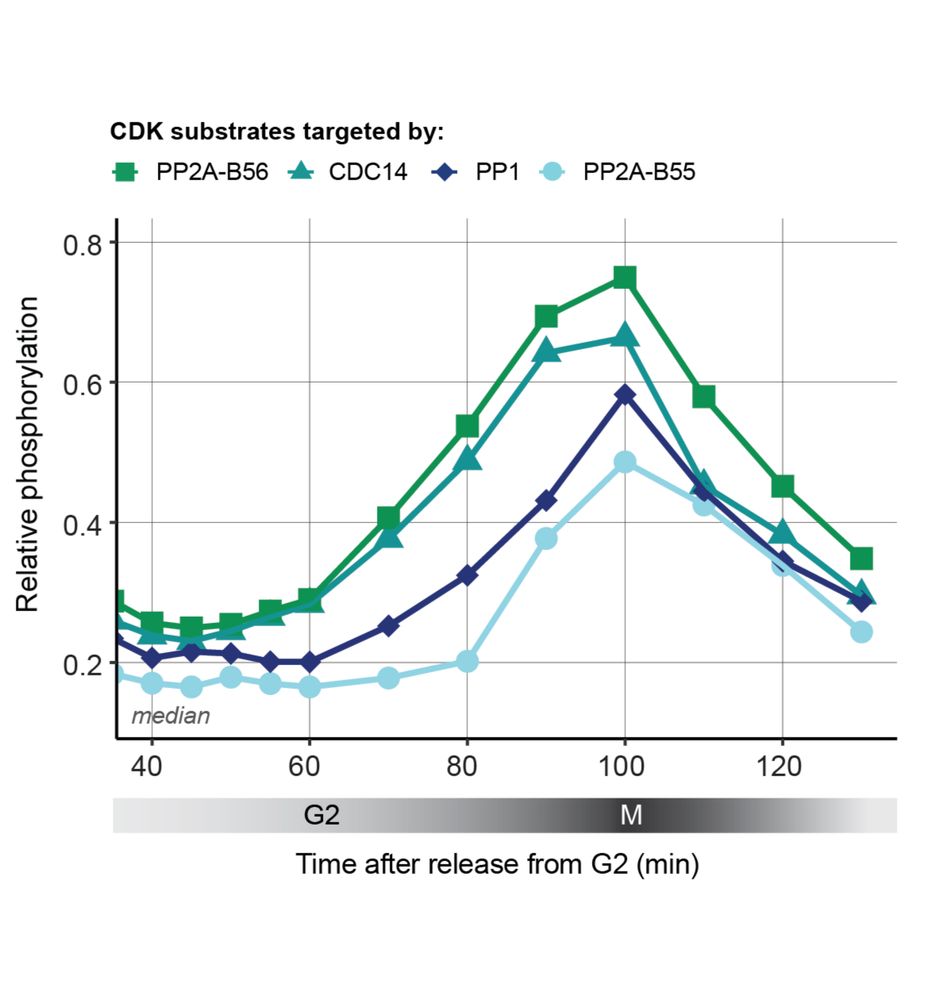
Phosphoproteomic timecourse showed that CDK substrates targeted by CDC14 and PP2A-B56 are, on average, net phosphorylated first, followed by CDK substrates targeted by PP1 and then PP2A-B55.
We also identified 3,000+ Ppase substrates phosphorylated by other kinases (not CDK). The identified Ppase motifs were similar to those identified in other eukaryotes, indicating conservation of Ppase substrate specificity.
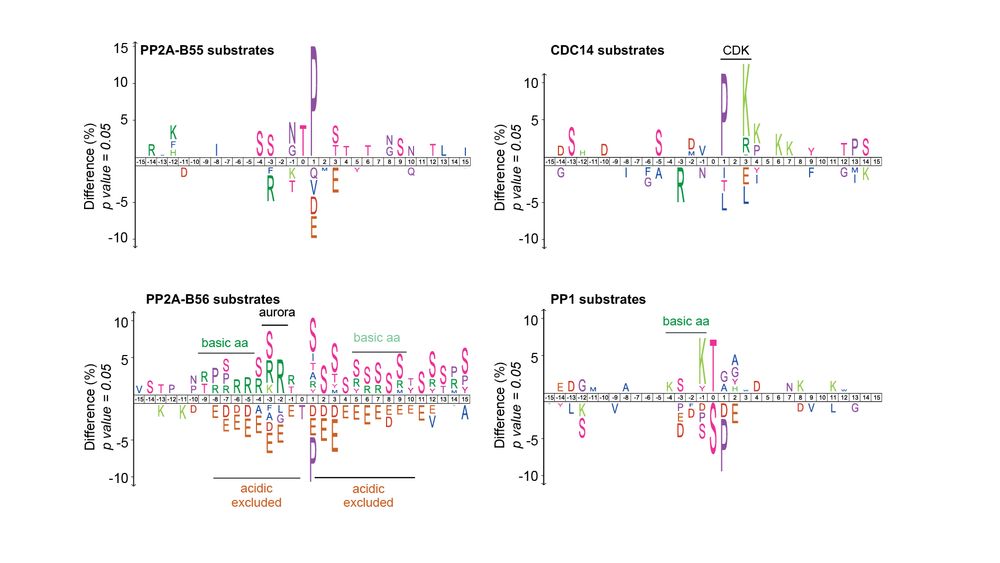
We also identified 3,000+ Ppase substrates phosphorylated by other kinases (not CDK). The identified Ppase motifs were similar to those identified in other eukaryotes, indicating conservation of Ppase substrate specificity.
• PP2A-B55, B56, CDC14 & PP1 targeted specific subgroups of CDK sites
• <5% of sites were targeted by multiple Ppases
• Ppases had different preferences for amino acids
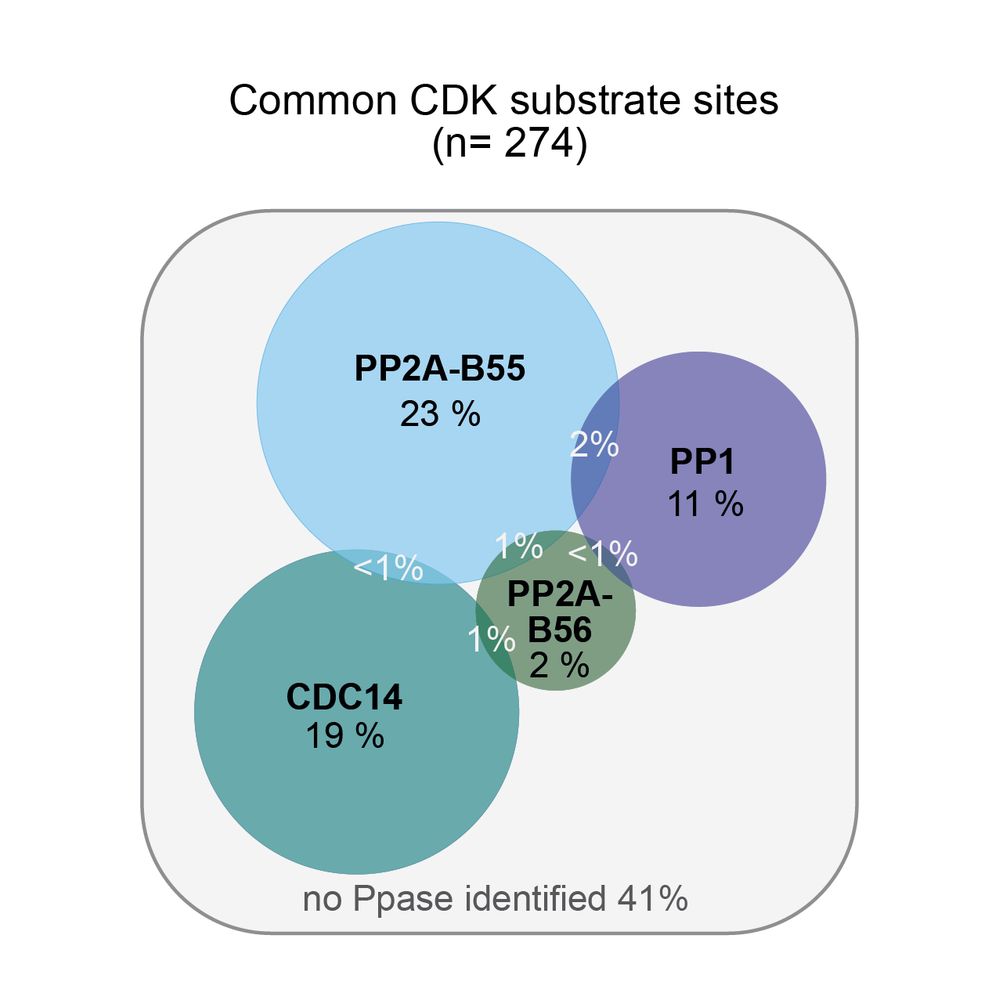
• PP2A-B55, B56, CDC14 & PP1 targeted specific subgroups of CDK sites
• <5% of sites were targeted by multiple Ppases
• Ppases had different preferences for amino acids
We used phosphoproteomics: inhibiting CDK with & without Ppase and tracking dephosphorylation. We classified a site as a Ppase substrate if it was dephosphorylated substantially slower in the absence of the Ppase.
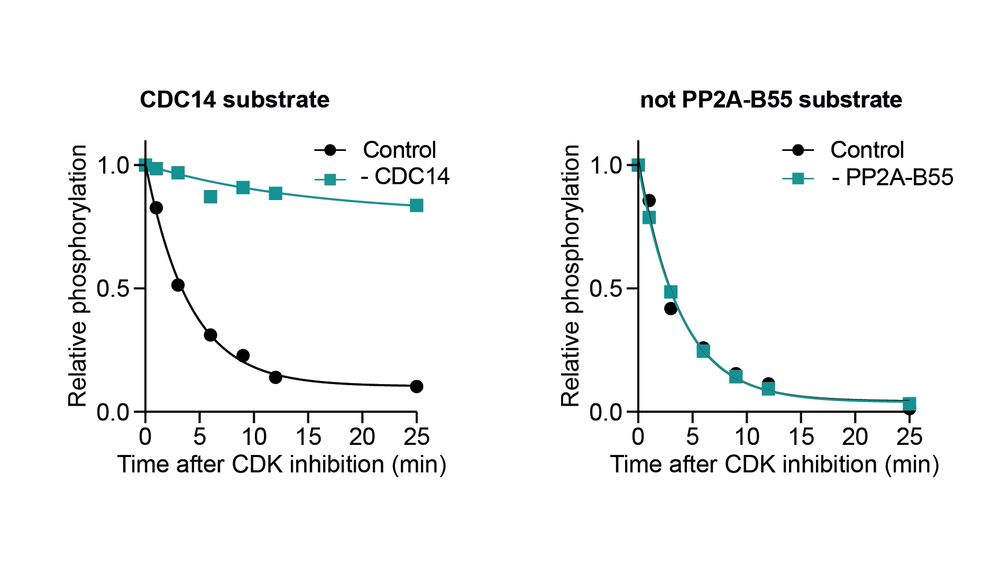
We used phosphoproteomics: inhibiting CDK with & without Ppase and tracking dephosphorylation. We classified a site as a Ppase substrate if it was dephosphorylated substantially slower in the absence of the Ppase.
How do CDK-opposing Ppases (PP2A-B55, PP2A-B56, PP1 & CDC14) together influence the phosphorylation timing of CDK substrates?
To answer this, we first needed to identify which CDK substrate is targeted by which Ppase.
How do CDK-opposing Ppases (PP2A-B55, PP2A-B56, PP1 & CDC14) together influence the phosphorylation timing of CDK substrates?
To answer this, we first needed to identify which CDK substrate is targeted by which Ppase.
• Important at mitotic exit to dephosphorylate CDK substrates
• Ppases are also active during interphase ➡️ could affect phosphorylation timing
• But for the majority of CDK substrate sites, it is not known which Ppase targets it
• Important at mitotic exit to dephosphorylate CDK substrates
• Ppases are also active during interphase ➡️ could affect phosphorylation timing
• But for the majority of CDK substrate sites, it is not known which Ppase targets it
Hundreds of proteins need to be phosphorylated at the right time by cyclin-dependent kinases (CDKs) to order cell cycle events. These phosphorylations are opposed by phosphatases (Ppases), such as PP2A-B55, PP2A-B56, PP1 & CDC14.
Hundreds of proteins need to be phosphorylated at the right time by cyclin-dependent kinases (CDKs) to order cell cycle events. These phosphorylations are opposed by phosphatases (Ppases), such as PP2A-B55, PP2A-B56, PP1 & CDC14.

