
Teif lab
@teiflab.bsky.social
Teif lab at the University of Essex. We work on gene regulation in chromatin and applications to liquid biopsies, using approaches of genomics, biophysics, bioinformatics & AI. Our focus is nucleosomics, TF binding, CTCF, cfDNA. https://generegulation.org
Bercovich et al., 2025. IceQream: Quantitative chromosome accessibility analysis using physical TF models www.nature.com/articles/s41...
▶️spatially integrates sequences and localises them relative to the target locus
▶️infers effective TF concentrations
▶️supplements with pairwise TF interactions
▶️spatially integrates sequences and localises them relative to the target locus
▶️infers effective TF concentrations
▶️supplements with pairwise TF interactions

October 26, 2025 at 2:25 PM
Bercovich et al., 2025. IceQream: Quantitative chromosome accessibility analysis using physical TF models www.nature.com/articles/s41...
▶️spatially integrates sequences and localises them relative to the target locus
▶️infers effective TF concentrations
▶️supplements with pairwise TF interactions
▶️spatially integrates sequences and localises them relative to the target locus
▶️infers effective TF concentrations
▶️supplements with pairwise TF interactions
It seems that it was not introduced by Google Scholar. It is a third-party add-on
project.iith.ac.in/sharmaglab/g...
addons.mozilla.org/en-US/firefo...
project.iith.ac.in/sharmaglab/g...
addons.mozilla.org/en-US/firefo...

October 26, 2025 at 10:56 AM
It seems that it was not introduced by Google Scholar. It is a third-party add-on
project.iith.ac.in/sharmaglab/g...
addons.mozilla.org/en-US/firefo...
project.iith.ac.in/sharmaglab/g...
addons.mozilla.org/en-US/firefo...
This is consistent with our observations, where in many cases statistically significant (and biologically informative) differences that occur between different states are just about 1-2bp or few bps that do not round to 5 (e.g. figure below from Piroeva et al., 2023) genome.cshlp.org/content/33/1...

October 7, 2025 at 9:43 AM
This is consistent with our observations, where in many cases statistically significant (and biologically informative) differences that occur between different states are just about 1-2bp or few bps that do not round to 5 (e.g. figure below from Piroeva et al., 2023) genome.cshlp.org/content/33/1...
When NRL is defined as a single genome-average value, it is usually not expected to be quantized. For example, this distribution of linker sizes from Voong et al, 2006 shows 10-bp quantization of preferred sizes, but the average of this distribution is expected to be a non-quantized value.

October 7, 2025 at 9:35 AM
When NRL is defined as a single genome-average value, it is usually not expected to be quantized. For example, this distribution of linker sizes from Voong et al, 2006 shows 10-bp quantization of preferred sizes, but the average of this distribution is expected to be a non-quantized value.
IT as a whole may be moved just about 100% up on this graph, but a sub-sector devoted to AI moved about 1000% up, so it might be interesting to repeat that earnings vs valuations analysis specifically for AI companies.

October 6, 2025 at 11:24 AM
IT as a whole may be moved just about 100% up on this graph, but a sub-sector devoted to AI moved about 1000% up, so it might be interesting to repeat that earnings vs valuations analysis specifically for AI companies.
Interesting paper from Gernot Längst & Co. They calculated nucleosome repeat length in Plasmodium falciparum, the malaria-causing parasite, and showed that it follows a smooth continuous function of time (the numbers on the X axis below are hours post-invasion) www.biorxiv.org/content/10.1...

October 4, 2025 at 7:01 PM
Interesting paper from Gernot Längst & Co. They calculated nucleosome repeat length in Plasmodium falciparum, the malaria-causing parasite, and showed that it follows a smooth continuous function of time (the numbers on the X axis below are hours post-invasion) www.biorxiv.org/content/10.1...
Polletti et al, 2025. Control of myeloid lineage fidelity and response to stimuli by ISWI-enforced nucleosome phasing www.cell.com/immunity/abs...
▶️PU.1 can form strongly phased nucleosome arrays similar to CTCF
▶️PU.1 can form strongly phased nucleosome arrays similar to CTCF

September 28, 2025 at 1:29 PM
Polletti et al, 2025. Control of myeloid lineage fidelity and response to stimuli by ISWI-enforced nucleosome phasing www.cell.com/immunity/abs...
▶️PU.1 can form strongly phased nucleosome arrays similar to CTCF
▶️PU.1 can form strongly phased nucleosome arrays similar to CTCF
Aging by the clock and yet without a program [perspective by David Meyer, Alexei Maklakov & Björn Schumacher] www.nature.com/articles/s43...



September 19, 2025 at 10:45 AM
Aging by the clock and yet without a program [perspective by David Meyer, Alexei Maklakov & Björn Schumacher] www.nature.com/articles/s43...
Hsu et al., 2025. MNase stratification reveals heterogeneous 5hmC in naive B cells www.biorxiv.org/content/10.1...

September 16, 2025 at 8:21 AM
Hsu et al., 2025. MNase stratification reveals heterogeneous 5hmC in naive B cells www.biorxiv.org/content/10.1...
my target price remains 200 for this one

September 8, 2025 at 7:05 PM
my target price remains 200 for this one
Contributions of local structural and energetic features of DNA to large-scale genomic organization [perspective by Wilma Olson & Co) www.sciencedirect.com/science/arti...




August 23, 2025 at 11:33 AM
Contributions of local structural and energetic features of DNA to large-scale genomic organization [perspective by Wilma Olson & Co) www.sciencedirect.com/science/arti...
Saunders et al., 2025. HMGB1 deforms nucleosomal DNA to generate a dynamic chromatin environment counteracting the effects of linker histone www.science.org/doi/full/10....
▶️HMGB1 sites are away from where H1 binds
▶️HMGB1 increases nucleosomal DNA accessibility without displacing H1
▶️HMGB1 sites are away from where H1 binds
▶️HMGB1 increases nucleosomal DNA accessibility without displacing H1

August 19, 2025 at 9:43 AM
Saunders et al., 2025. HMGB1 deforms nucleosomal DNA to generate a dynamic chromatin environment counteracting the effects of linker histone www.science.org/doi/full/10....
▶️HMGB1 sites are away from where H1 binds
▶️HMGB1 increases nucleosomal DNA accessibility without displacing H1
▶️HMGB1 sites are away from where H1 binds
▶️HMGB1 increases nucleosomal DNA accessibility without displacing H1
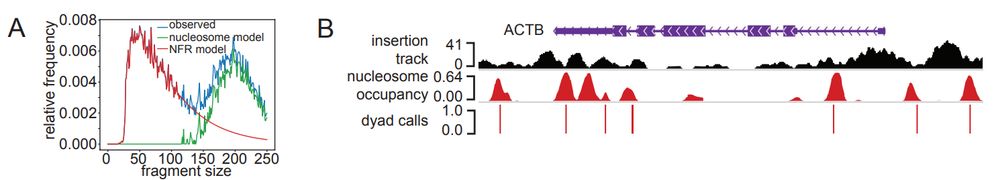
Liu et al., 2025. An automated ATAC-seq method reveals sequence determinants of transcription factor dose response in the open chromatin www.biorxiv.org/content/10.1...
▶️RoboATAC - automated ATAC-seq
▶️Deep learning models: motif orientation, spacing, flanking bases
▶️Nucleosome positioning analysis
▶️RoboATAC - automated ATAC-seq
▶️Deep learning models: motif orientation, spacing, flanking bases
▶️Nucleosome positioning analysis
July 27, 2025 at 12:31 PM
Liu et al., 2025. An automated ATAC-seq method reveals sequence determinants of transcription factor dose response in the open chromatin www.biorxiv.org/content/10.1...
▶️RoboATAC - automated ATAC-seq
▶️Deep learning models: motif orientation, spacing, flanking bases
▶️Nucleosome positioning analysis
▶️RoboATAC - automated ATAC-seq
▶️Deep learning models: motif orientation, spacing, flanking bases
▶️Nucleosome positioning analysis
Wang et al, 2025. Genome-Wide Investigation of Transcription Factor Occupancy and Dynamics Using cFOOT-seq www.biorxiv.org/content/10.1...

July 22, 2025 at 8:55 AM
Wang et al, 2025. Genome-Wide Investigation of Transcription Factor Occupancy and Dynamics Using cFOOT-seq www.biorxiv.org/content/10.1...
This graph shows no correlation woth GBP/USD

July 10, 2025 at 10:57 AM
This graph shows no correlation woth GBP/USD
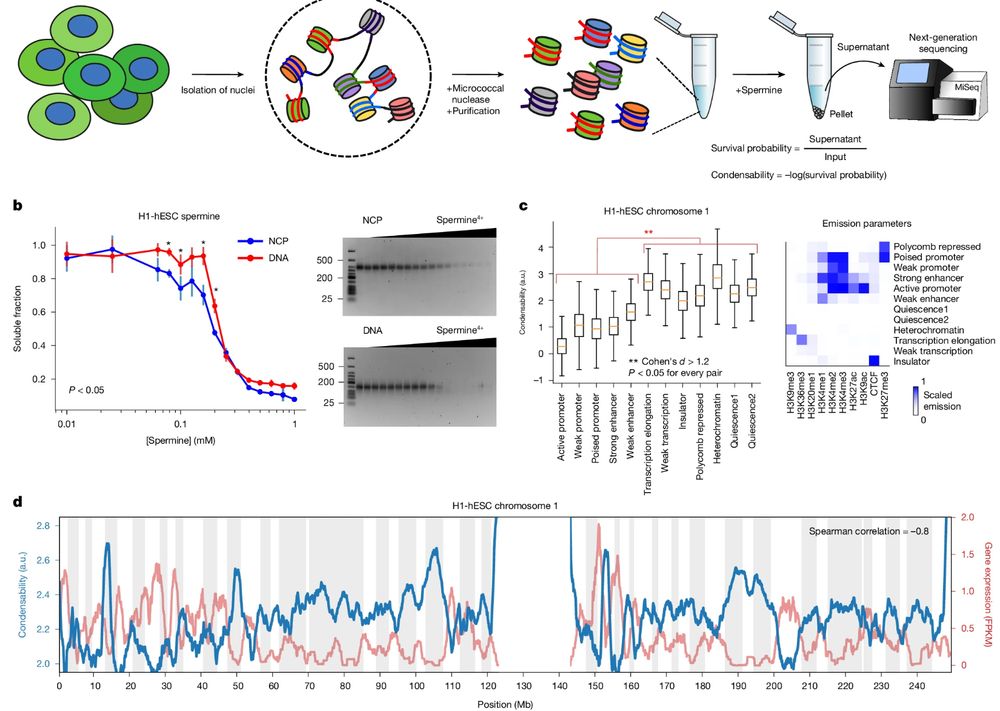
Native nucleosomes intrinsically encode genome organization principles www.nature.com/articles/s41...
▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
May 10, 2025 at 7:42 PM
Native nucleosomes intrinsically encode genome organization principles www.nature.com/articles/s41...
▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
Kable et al., 2025. Compromised 2-start zigzag chromatin folding in immature mouse retina cells driven by irregularly spaced nucleosomes with short DNA linkers www.biorxiv.org/content/10.1...
▶️In mature retina cells, the fraction of short-linker nucleosomes is lower -> stronger chromatin compaction.
▶️In mature retina cells, the fraction of short-linker nucleosomes is lower -> stronger chromatin compaction.

May 5, 2025 at 12:52 PM
Kable et al., 2025. Compromised 2-start zigzag chromatin folding in immature mouse retina cells driven by irregularly spaced nucleosomes with short DNA linkers www.biorxiv.org/content/10.1...
▶️In mature retina cells, the fraction of short-linker nucleosomes is lower -> stronger chromatin compaction.
▶️In mature retina cells, the fraction of short-linker nucleosomes is lower -> stronger chromatin compaction.
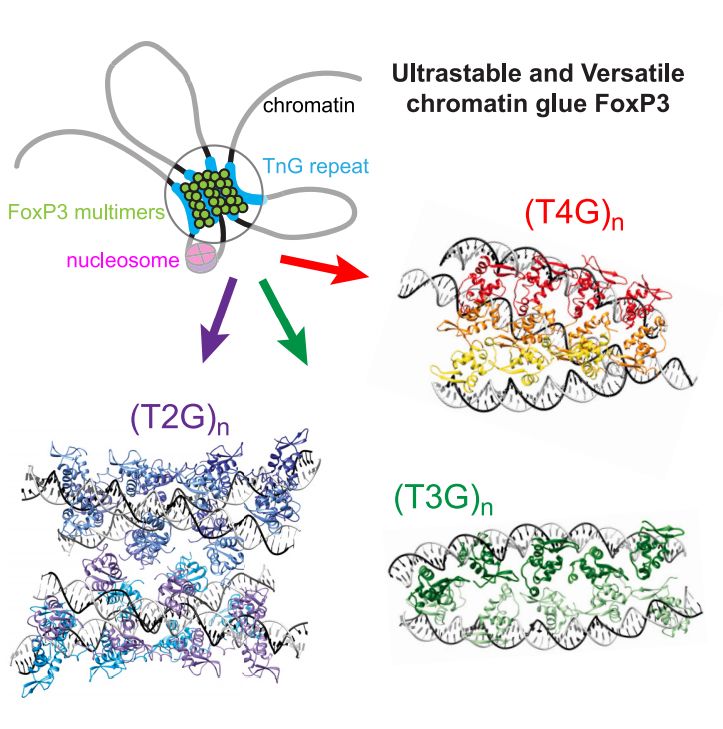
Ultrastable and versatile multimeric ensembles of FoxP3 on microsatellites www.cell.com/molecular-ce...
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
April 5, 2025 at 9:54 PM
Ultrastable and versatile multimeric ensembles of FoxP3 on microsatellites www.cell.com/molecular-ce...
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
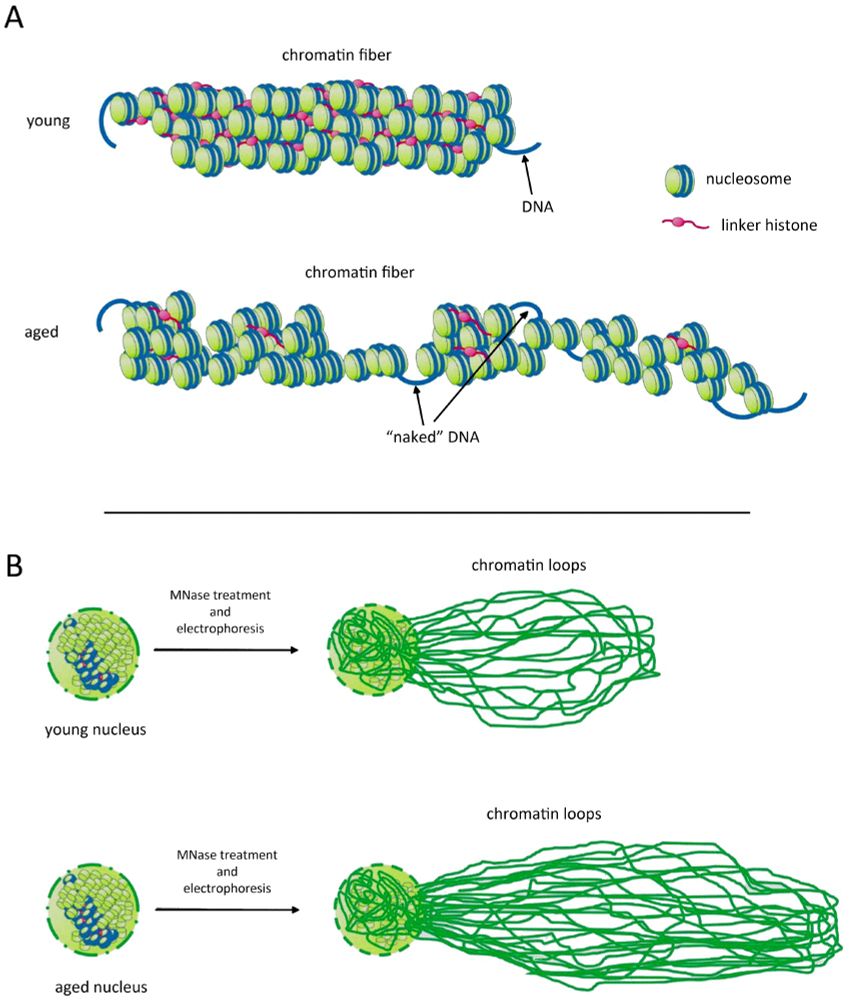
Miloshev et al., 2025, Linker Histones Maintain Genome Stability and Drive the Process of Cellular Ageing [review] www.imrpress.com/journal/FBL/...
Interesting figure: we did observe NRL increase with ageing (onlinelibrary.wiley.com/doi/10.1111/...). Not sure about about loop sizes, to be checked
Interesting figure: we did observe NRL increase with ageing (onlinelibrary.wiley.com/doi/10.1111/...). Not sure about about loop sizes, to be checked
April 1, 2025 at 10:08 AM
Miloshev et al., 2025, Linker Histones Maintain Genome Stability and Drive the Process of Cellular Ageing [review] www.imrpress.com/journal/FBL/...
Interesting figure: we did observe NRL increase with ageing (onlinelibrary.wiley.com/doi/10.1111/...). Not sure about about loop sizes, to be checked
Interesting figure: we did observe NRL increase with ageing (onlinelibrary.wiley.com/doi/10.1111/...). Not sure about about loop sizes, to be checked
Evolutionary divergence in CTCF-mediated chromatin topology drives transcriptional innovation in humans www.nature.com/articles/s41...
Human vs macaque:
▶️2133 gained (2.7%), 2418 lost (3.1%) CTCF loops in B-cells
▶️5873 gained (7.3%), 6708 lost (8.3%) in neurons
▶️implications for ASD and immunity
Human vs macaque:
▶️2133 gained (2.7%), 2418 lost (3.1%) CTCF loops in B-cells
▶️5873 gained (7.3%), 6708 lost (8.3%) in neurons
▶️implications for ASD and immunity

March 30, 2025 at 2:19 PM
Evolutionary divergence in CTCF-mediated chromatin topology drives transcriptional innovation in humans www.nature.com/articles/s41...
Human vs macaque:
▶️2133 gained (2.7%), 2418 lost (3.1%) CTCF loops in B-cells
▶️5873 gained (7.3%), 6708 lost (8.3%) in neurons
▶️implications for ASD and immunity
Human vs macaque:
▶️2133 gained (2.7%), 2418 lost (3.1%) CTCF loops in B-cells
▶️5873 gained (7.3%), 6708 lost (8.3%) in neurons
▶️implications for ASD and immunity
Do et al., 2025. Binding domain mutations provide insight into CTCF’s relationship with chromatin and its contribution to gene regulation www.cell.com/cell-genomic...

March 23, 2025 at 7:55 AM
Do et al., 2025. Binding domain mutations provide insight into CTCF’s relationship with chromatin and its contribution to gene regulation www.cell.com/cell-genomic...






