@SEMC_NYSBC. Co-founder and CEO of http://OpenProtein.AI. Opinions are my own.
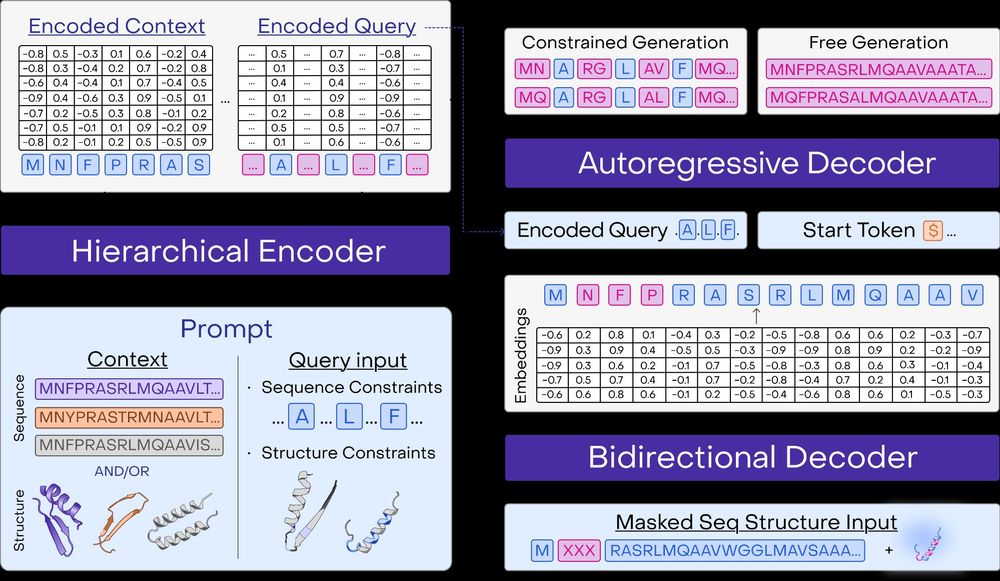
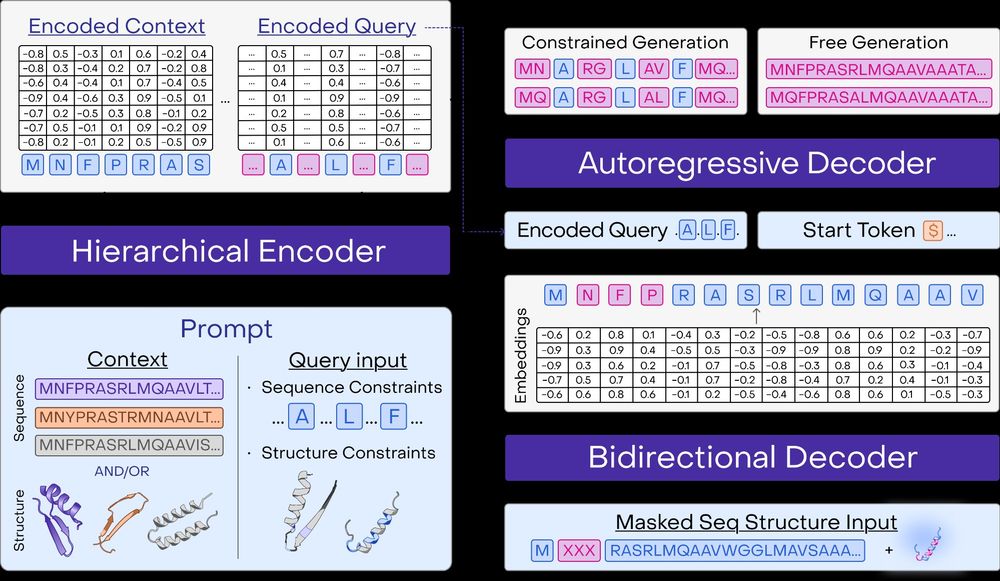
and install the python client to get started: github.com/OpenProteinA...

and install the python client to get started: github.com/OpenProteinA...
I can’t wait to see what the community can do with these models! 13/13
I can’t wait to see what the community can do with these models! 13/13
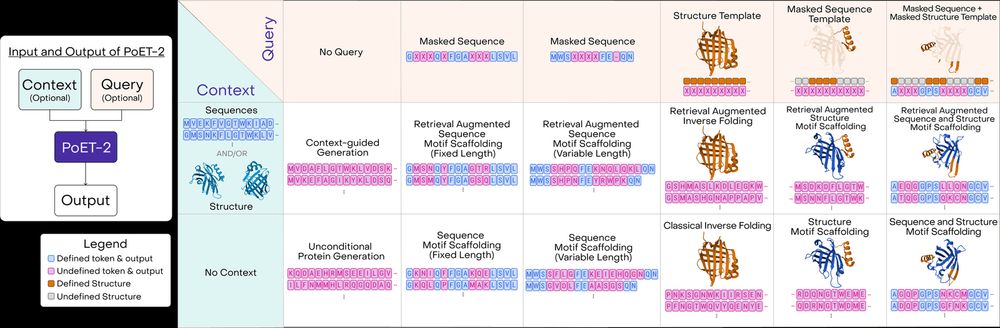
12/13
12/13
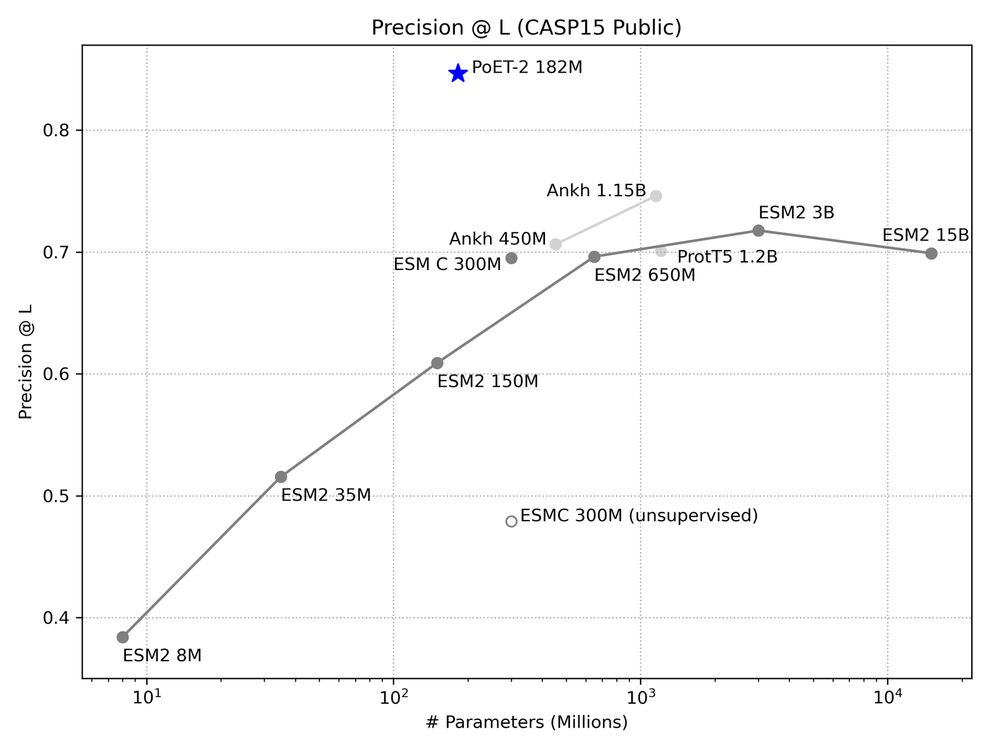
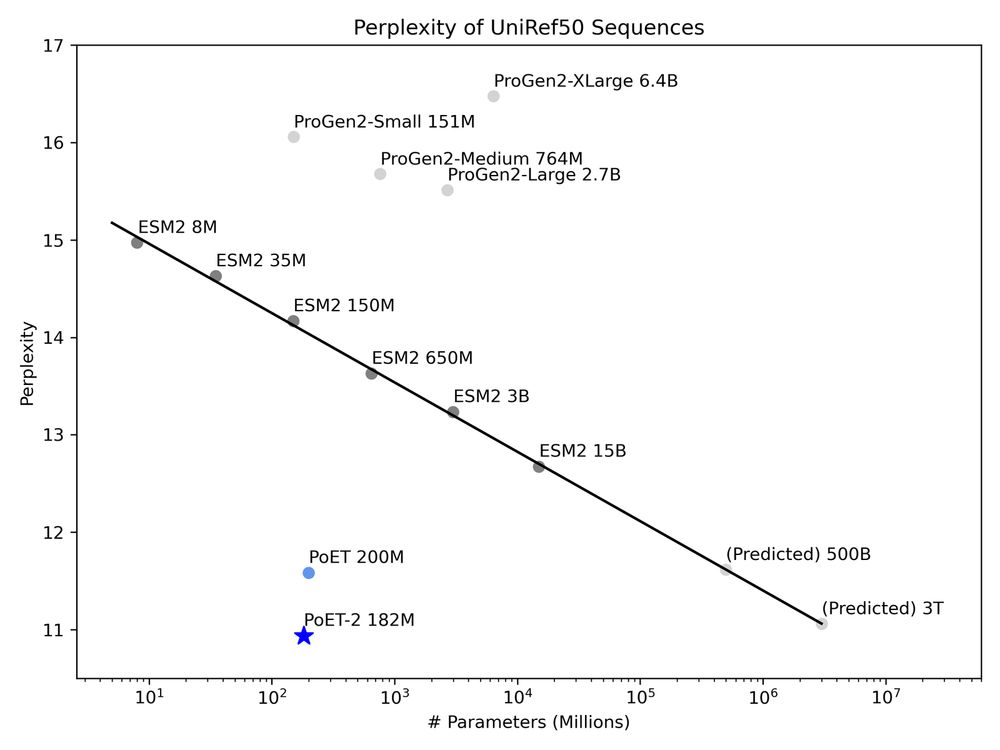
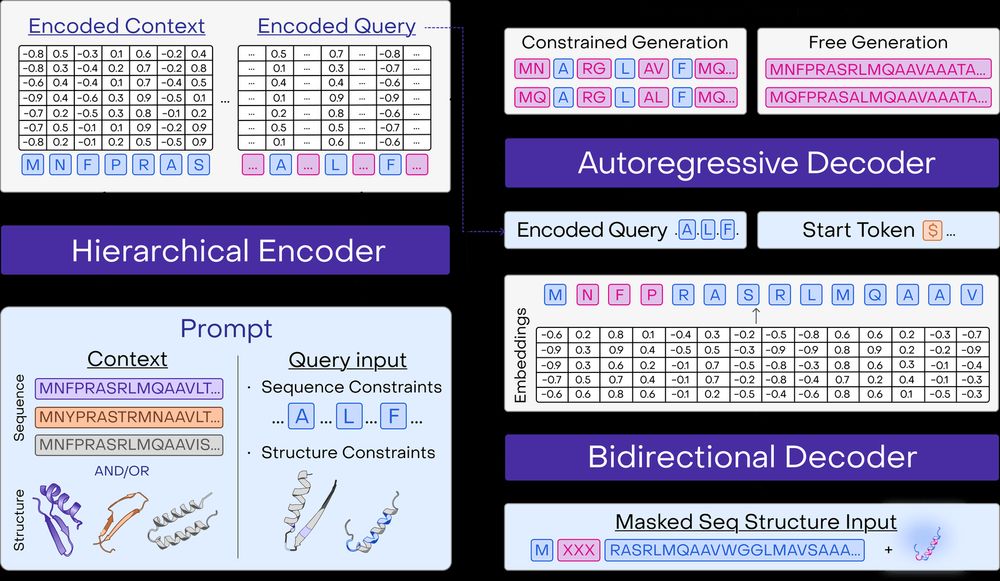
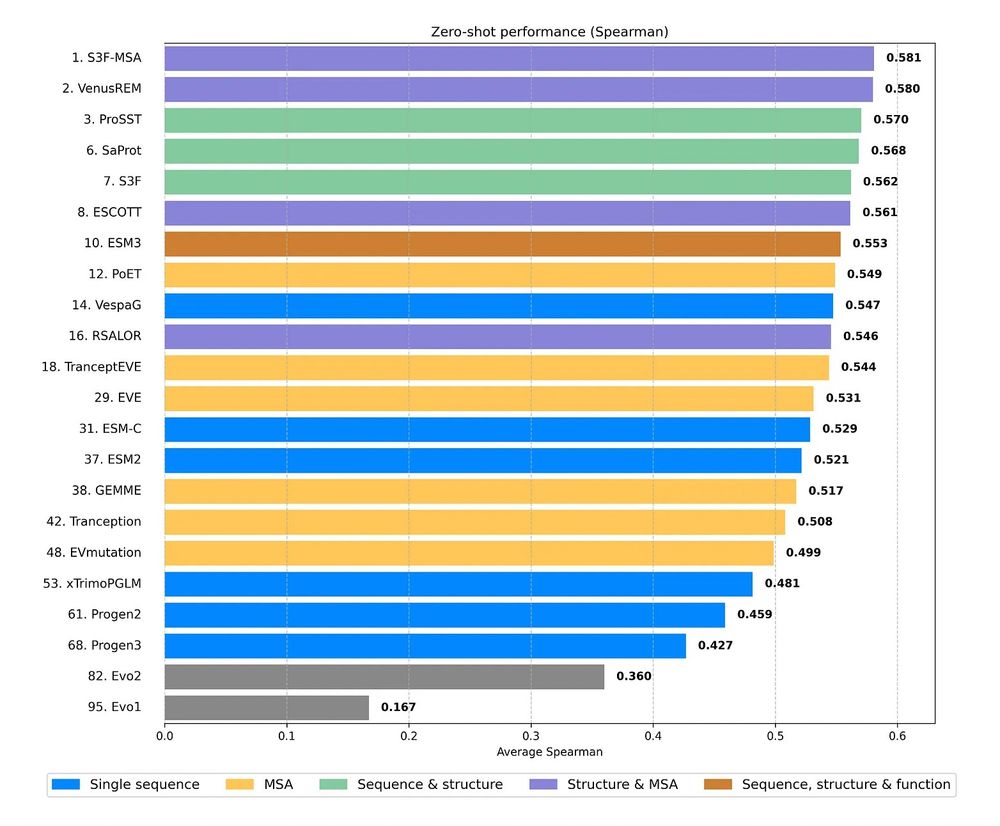
* Improves sequence and structure understanding
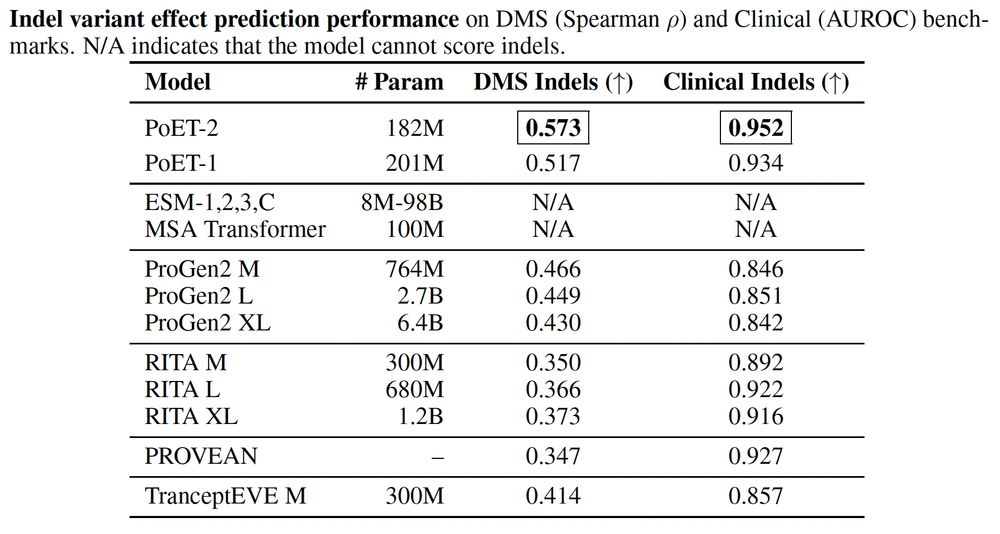
* Accurate zero-shot function prediction, especially for insertions and deletions
* 30x less data needed for transfer learning
8/13
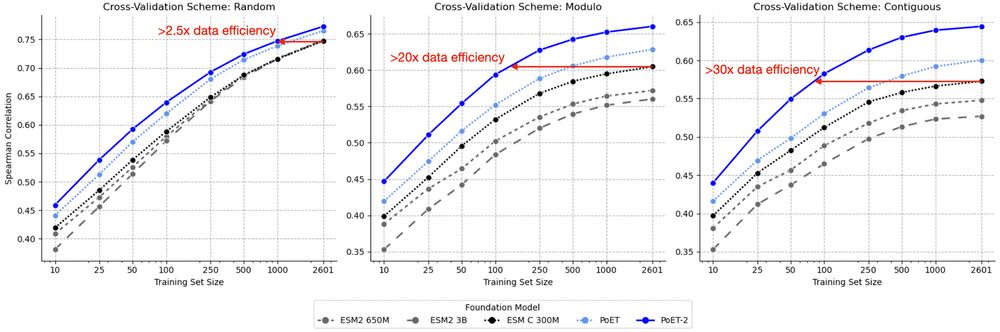
* Improves sequence and structure understanding
* Accurate zero-shot function prediction, especially for insertions and deletions
* 30x less data needed for transfer learning
8/13