
Tauras Vilgalys
@taurvil.bsky.social
Postdoc at UChicago. Evolutionary functional genomics, focusing on how humans and other primates respond to infectious disease. Former grad student @EvAnthDuke. 🧪
Many thanks to @jennytung.bsky.social, @afogel29.bsky.social, Jordan Anderson, and all the other members of the @amboselibaboonrp.bsky.social who made this work possible!
August 15, 2025 at 4:23 PM
Many thanks to @jennytung.bsky.social, @afogel29.bsky.social, Jordan Anderson, and all the other members of the @amboselibaboonrp.bsky.social who made this work possible!
Together, our results suggest that the genetic architecture of DNA methylation is conserved between closely related species. Cis-genetic effects on gene regulation may remain stable for over a million years, which has implications for how we understand modern and archaic human variants.
August 15, 2025 at 4:23 PM
Together, our results suggest that the genetic architecture of DNA methylation is conserved between closely related species. Cis-genetic effects on gene regulation may remain stable for over a million years, which has implications for how we understand modern and archaic human variants.
We also show that ancestry effects within Amboseli recapitulates differences between the parent species. These results support a genetic basis for many interspecific methylation differences, despite environmental variation when individuals are sampled from distinct populations.

August 15, 2025 at 4:23 PM
We also show that ancestry effects within Amboseli recapitulates differences between the parent species. These results support a genetic basis for many interspecific methylation differences, despite environmental variation when individuals are sampled from distinct populations.
By changing the allele frequency, admixture introduces new regulatory variation - including at sites that would be otherwise fixed in the population!
August 15, 2025 at 4:23 PM
By changing the allele frequency, admixture introduces new regulatory variation - including at sites that would be otherwise fixed in the population!
Local genetic ancestry frequently predicts DNA methylation levels, and these differences can be predicted using the allele frequency at cis-acting genetic variants (meQTL).

August 15, 2025 at 4:23 PM
Local genetic ancestry frequently predicts DNA methylation levels, and these differences can be predicted using the allele frequency at cis-acting genetic variants (meQTL).
There's some nice work on how the power of selection scans differs based on the time of selection. I can't remember where that figure was published though
May 8, 2025 at 7:22 PM
There's some nice work on how the power of selection scans differs based on the time of selection. I can't remember where that figure was published though
Thanks Jun, I couldn't agree more!
May 5, 2025 at 7:55 PM
Thanks Jun, I couldn't agree more!
Finally, I am immensely grateful to my mentors over the years (particularly @jennytung.bsky.social and Luis Barreiro), as well as all of the collaborators, lab members, and friends who have helped along this journey!
May 5, 2025 at 4:31 PM
Finally, I am immensely grateful to my mentors over the years (particularly @jennytung.bsky.social and Luis Barreiro), as well as all of the collaborators, lab members, and friends who have helped along this journey!
I will be recruiting students, postdocs, and technicians -- if you're interested in this work, please reach out to me!
May 5, 2025 at 4:31 PM
I will be recruiting students, postdocs, and technicians -- if you're interested in this work, please reach out to me!
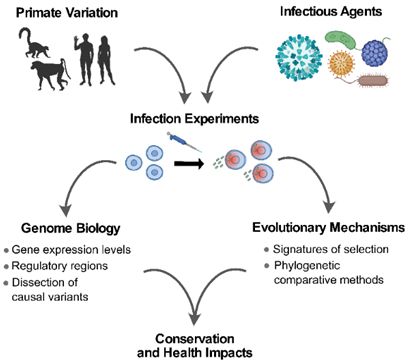
My lab will study how genetic varation among primates shapes the response to environmental stimuli, focusing on the immune system. Ultimately, I hope to connect genetic variation to immune phenotypes and identify the ecological and evolutionary drivers of immune variation.
May 5, 2025 at 4:31 PM
My lab will study how genetic varation among primates shapes the response to environmental stimuli, focusing on the immune system. Ultimately, I hope to connect genetic variation to immune phenotypes and identify the ecological and evolutionary drivers of immune variation.

