Stanley E. Strawbridge
@stanleystrawbridge.bsky.social
Physics of Life & Quantitative Biology Fellow
Group Leader at University of Sheffield
Stem Cells // Development // Math(s) // ML
WV -> Ox -> Cam -> Shef
strawbridgelab.com
Group Leader at University of Sheffield
Stem Cells // Development // Math(s) // ML
WV -> Ox -> Cam -> Shef
strawbridgelab.com
Do you make shipments?! I’d love this for the lab!
July 1, 2025 at 10:27 PM
Do you make shipments?! I’d love this for the lab!
Finally, a career highlight, we made it into @cellysally.bsky.social 's #PaperThemeTune s:
bsky.app/profile/cell...
[8/8]
bsky.app/profile/cell...
[8/8]
Strawbridge et al use genetic tricks and mathematical modelling to work out how donor ES cells displace host cells to build chimeric embryos
Do they politely converse with them via chemical signalling? Or do they just rudely push them out?
#PaperThemeTune “Push it out” by the Beta Band
#DevBio
Do they politely converse with them via chemical signalling? Or do they just rudely push them out?
#PaperThemeTune “Push it out” by the Beta Band
#DevBio
Donor embryonic stem cells impede host epiblast specification in 8-cell stage chimeras by crowding and FGF4 signalling https://www.biorxiv.org/content/10.1101/2024.11.08.622647v1
June 24, 2025 at 3:01 PM
Finally, a career highlight, we made it into @cellysally.bsky.social 's #PaperThemeTune s:
bsky.app/profile/cell...
[8/8]
bsky.app/profile/cell...
[8/8]
Also, check out the behind-the-scenes interview on my path in science, a eureka moment, and what I do when I choose to leave the lab:
🎙️ doi.org/10.1242/dev....
[7/8]
🎙️ doi.org/10.1242/dev....
[7/8]
June 24, 2025 at 3:01 PM
Also, check out the behind-the-scenes interview on my path in science, a eureka moment, and what I do when I choose to leave the lab:
🎙️ doi.org/10.1242/dev....
[7/8]
🎙️ doi.org/10.1242/dev....
[7/8]
June 24, 2025 at 3:01 PM
Thanks to co-authors & host institutions:
@scicambridge.bsky.social @pdncambridge.bsky.social @cam.ac.uk @drn-sheffield.bsky.social @sheffielduni.bsky.social @uoe-igc.bsky.social @edinburgh-uni.bsky.social @mskcancercenter.bsky.social @uniheidelberg.bsky.social @univparissaclay.bsky.social
[5/8]
@scicambridge.bsky.social @pdncambridge.bsky.social @cam.ac.uk @drn-sheffield.bsky.social @sheffielduni.bsky.social @uoe-igc.bsky.social @edinburgh-uni.bsky.social @mskcancercenter.bsky.social @uniheidelberg.bsky.social @univparissaclay.bsky.social
[5/8]
June 24, 2025 at 3:01 PM
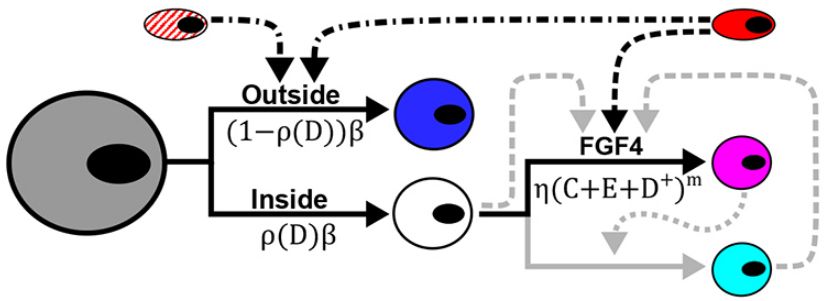
To test this, we built a quantitative model of fate specification in the embryo. It predicts a two-step mechanism:
1️⃣ Physical exclusion
2️⃣ FGF4-driven fate bias
And it even works at the level of individual embryos!
[4/8]
1️⃣ Physical exclusion
2️⃣ FGF4-driven fate bias
And it even works at the level of individual embryos!
[4/8]
June 24, 2025 at 3:01 PM
To test this, we built a quantitative model of fate specification in the embryo. It predicts a two-step mechanism:
1️⃣ Physical exclusion
2️⃣ FGF4-driven fate bias
And it even works at the level of individual embryos!
[4/8]
1️⃣ Physical exclusion
2️⃣ FGF4-driven fate bias
And it even works at the level of individual embryos!
[4/8]
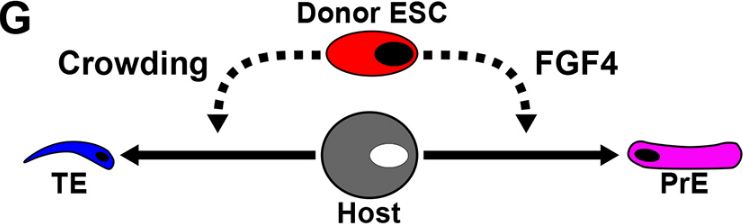
Even ESCs lacking FGF4 can push host cells into the trophectoderm, just by outcompeting them for space. But with FGF4, they also bias host cells away from contributing to the embryo.
➡️ Crowding first, then signalling.
[3/8]
➡️ Crowding first, then signalling.
[3/8]
June 24, 2025 at 3:01 PM
Even ESCs lacking FGF4 can push host cells into the trophectoderm, just by outcompeting them for space. But with FGF4, they also bias host cells away from contributing to the embryo.
➡️ Crowding first, then signalling.
[3/8]
➡️ Crowding first, then signalling.
[3/8]
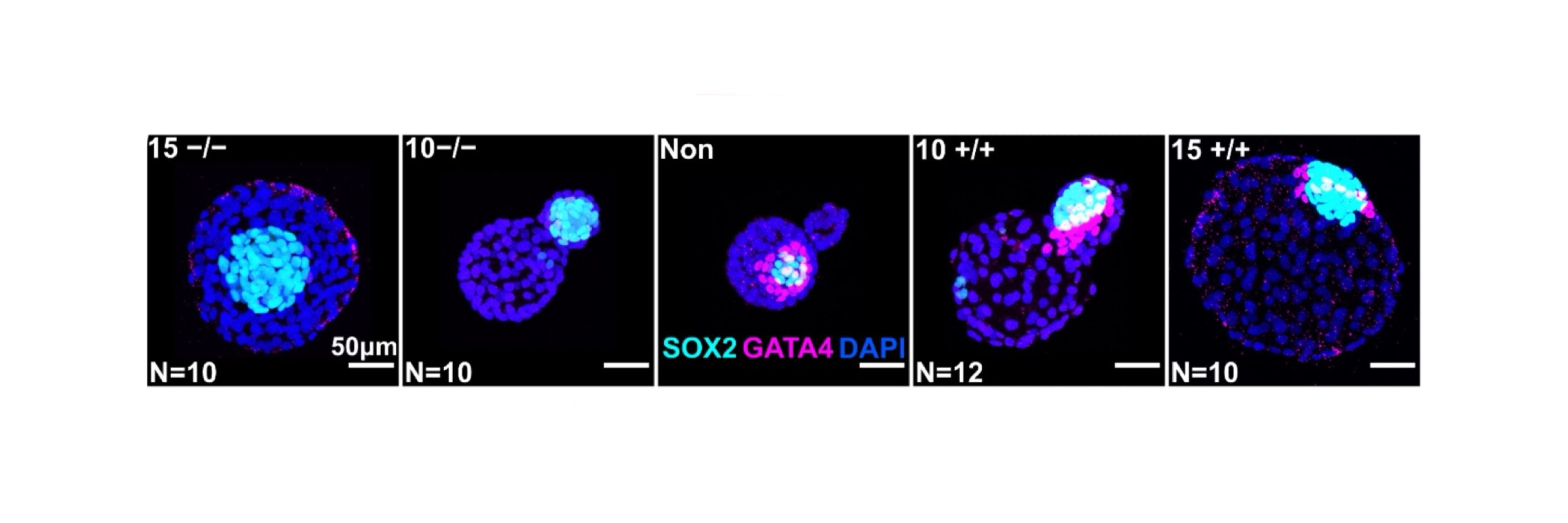
When injected into 8-cell embryos, ESCs displace host cells from the future embryo into extra-embryonic tissues. This happens via physical crowding and, if the donor cells produce FGF4, also via molecular signalling.
[2/8]
[2/8]
June 24, 2025 at 3:01 PM
When injected into 8-cell embryos, ESCs displace host cells from the future embryo into extra-embryonic tissues. This happens via physical crowding and, if the donor cells produce FGF4, also via molecular signalling.
[2/8]
[2/8]
I’ve been using my CoB insulated lunch bag for work ever since Biologist @ 100 in Liverpool! 🤓
June 12, 2025 at 11:37 AM
I’ve been using my CoB insulated lunch bag for work ever since Biologist @ 100 in Liverpool! 🤓
10/10 Thanks to:
#MRC
@wellcometrust.bsky.social
#SãoPauloResearchFoundation
#JapanSocietyforthePromotionofScience #JSPS
#UEHARAMemorialFoundation
#MRC
@wellcometrust.bsky.social
#SãoPauloResearchFoundation
#JapanSocietyforthePromotionofScience #JSPS
#UEHARAMemorialFoundation
June 4, 2025 at 6:17 PM
9/10 @drn-sheffield.bsky.social @sheffielduni.bsky.social @uoe-igc.bsky.social @edinburgh-uni.bsky.social @uniofaberdeen.bsky.social @uspoficial.bsky.social @lsiexeter.bsky.social @exeter.ac.uk
June 4, 2025 at 6:17 PM
8/10 Thank you to all the excellent people involved! (That I can find)
@lbates.bsky.social @tazami.bsky.social
Thanks to: @scicambridge.bsky.social @pdncambridge.bsky.social @babrahaminst.bsky.social @cambiochem.bsky.social @cam.ac.uk ...
@lbates.bsky.social @tazami.bsky.social
Thanks to: @scicambridge.bsky.social @pdncambridge.bsky.social @babrahaminst.bsky.social @cambiochem.bsky.social @cam.ac.uk ...
June 4, 2025 at 6:17 PM
8/10 Thank you to all the excellent people involved! (That I can find)
@lbates.bsky.social @tazami.bsky.social
Thanks to: @scicambridge.bsky.social @pdncambridge.bsky.social @babrahaminst.bsky.social @cambiochem.bsky.social @cam.ac.uk ...
@lbates.bsky.social @tazami.bsky.social
Thanks to: @scicambridge.bsky.social @pdncambridge.bsky.social @babrahaminst.bsky.social @cambiochem.bsky.social @cam.ac.uk ...
7/10 Somatic versatility
Capacitated cultures then generate neuroectoderm, paraxial mesoderm & definitive endoderm with textbook marker profiles—ready for downstream lineage studies.
Capacitated cultures then generate neuroectoderm, paraxial mesoderm & definitive endoderm with textbook marker profiles—ready for downstream lineage studies.
June 4, 2025 at 6:17 PM
7/10 Somatic versatility
Capacitated cultures then generate neuroectoderm, paraxial mesoderm & definitive endoderm with textbook marker profiles—ready for downstream lineage studies.
Capacitated cultures then generate neuroectoderm, paraxial mesoderm & definitive endoderm with textbook marker profiles—ready for downstream lineage studies.
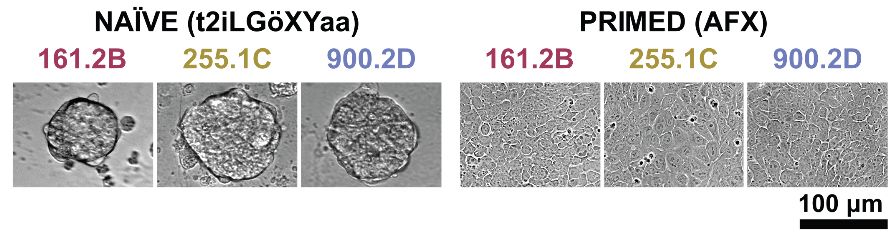
6/10 Forward Progression
10-day XAV capacitation guides naïve domes → primed flats—an in-vitro window on the pre- to post-implantation transition.
10-day XAV capacitation guides naïve domes → primed flats—an in-vitro window on the pre- to post-implantation transition.
June 4, 2025 at 6:17 PM
6/10 Forward Progression
10-day XAV capacitation guides naïve domes → primed flats—an in-vitro window on the pre- to post-implantation transition.
10-day XAV capacitation guides naïve domes → primed flats—an in-vitro window on the pre- to post-implantation transition.
5/10 Extra-embryonic power
PD03 + A83 + Y → GATA3⁺ trophoblast cysts.
LIF + ACTIVIN + CHIR → hypoblast/ExEn. Use them for blastoids or placenta work.
PD03 + A83 + Y → GATA3⁺ trophoblast cysts.
LIF + ACTIVIN + CHIR → hypoblast/ExEn. Use them for blastoids or placenta work.
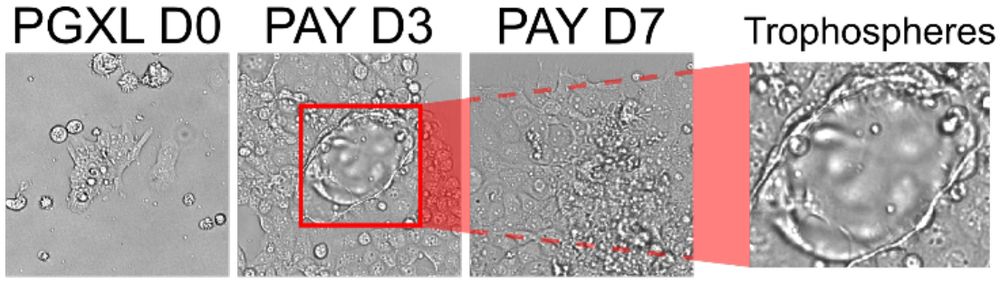
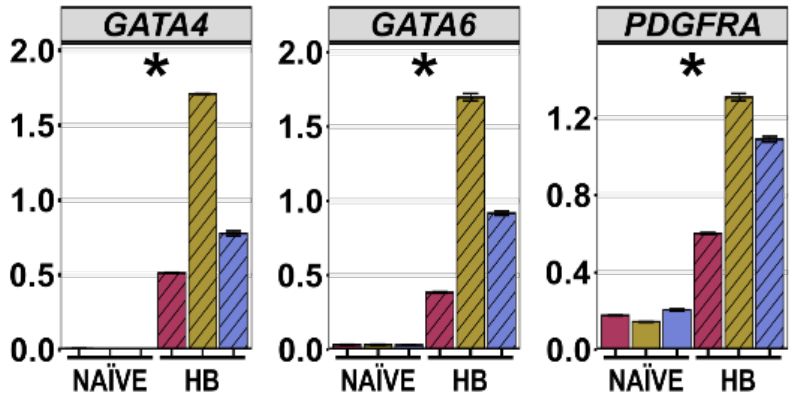
June 4, 2025 at 6:17 PM
5/10 Extra-embryonic power
PD03 + A83 + Y → GATA3⁺ trophoblast cysts.
LIF + ACTIVIN + CHIR → hypoblast/ExEn. Use them for blastoids or placenta work.
PD03 + A83 + Y → GATA3⁺ trophoblast cysts.
LIF + ACTIVIN + CHIR → hypoblast/ExEn. Use them for blastoids or placenta work.
4/10 Hallmark naïve identity
Lines keep the classic naïve signature, form domes in t2iLGöXYaa, and map to E5/6 epiblast by bulk RNA-seq.
Lines keep the classic naïve signature, form domes in t2iLGöXYaa, and map to E5/6 epiblast by bulk RNA-seq.
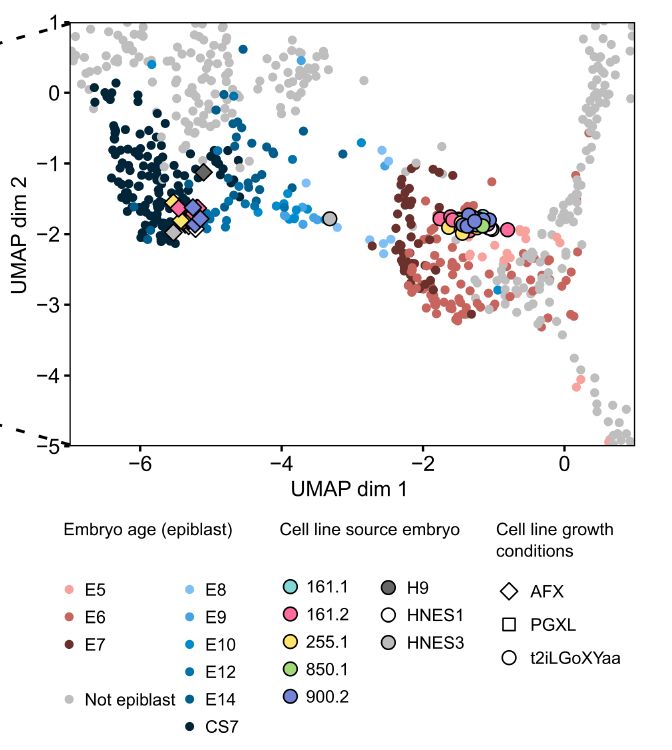
June 4, 2025 at 6:17 PM
4/10 Hallmark naïve identity
Lines keep the classic naïve signature, form domes in t2iLGöXYaa, and map to E5/6 epiblast by bulk RNA-seq.
Lines keep the classic naïve signature, form domes in t2iLGöXYaa, and map to E5/6 epiblast by bulk RNA-seq.
3/10 Mosaicism in a dish
Two triplet sets showcase natural variation:
• 161.2A/C share a Chr2q deletion, 161.2B does not.
• 255.1B/C carry Chr5 trisomy, 255.1A does not.
Study how karyotype skews lineage choice—no embryos needed.
Two triplet sets showcase natural variation:
• 161.2A/C share a Chr2q deletion, 161.2B does not.
• 255.1B/C carry Chr5 trisomy, 255.1A does not.
Study how karyotype skews lineage choice—no embryos needed.
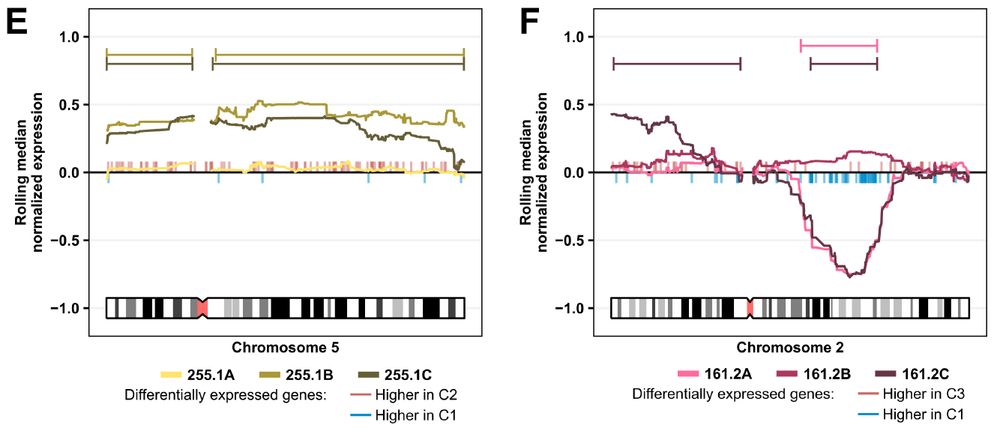
June 4, 2025 at 6:17 PM
3/10 Mosaicism in a dish
Two triplet sets showcase natural variation:
• 161.2A/C share a Chr2q deletion, 161.2B does not.
• 255.1B/C carry Chr5 trisomy, 255.1A does not.
Study how karyotype skews lineage choice—no embryos needed.
Two triplet sets showcase natural variation:
• 161.2A/C share a Chr2q deletion, 161.2B does not.
• 255.1B/C carry Chr5 trisomy, 255.1A does not.
Study how karyotype skews lineage choice—no embryos needed.
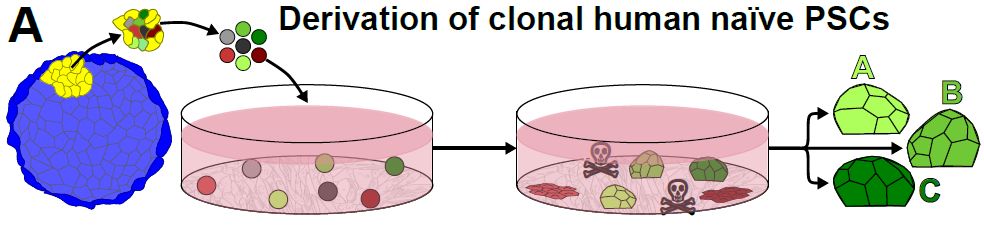
2/10 Multiple clones per embryo
Instead of one line per blastocyst, we expanded every dome-shaped colony. Result: single embryos now come with paired, triplet or quadruplet lines—perfect for side-by-side comparisons.
Instead of one line per blastocyst, we expanded every dome-shaped colony. Result: single embryos now come with paired, triplet or quadruplet lines—perfect for side-by-side comparisons.
June 4, 2025 at 6:17 PM
2/10 Multiple clones per embryo
Instead of one line per blastocyst, we expanded every dome-shaped colony. Result: single embryos now come with paired, triplet or quadruplet lines—perfect for side-by-side comparisons.
Instead of one line per blastocyst, we expanded every dome-shaped colony. Result: single embryos now come with paired, triplet or quadruplet lines—perfect for side-by-side comparisons.

