
Current: Evolutionary Virologist based at the University of Tokyo 🇯🇵
💻🦠🏳️🌈
Slight change to my office window view from Tokyo Tower🗼 to the Tour Eiffel. 🇫🇷

Slight change to my office window view from Tokyo Tower🗼 to the Tour Eiffel. 🇫🇷









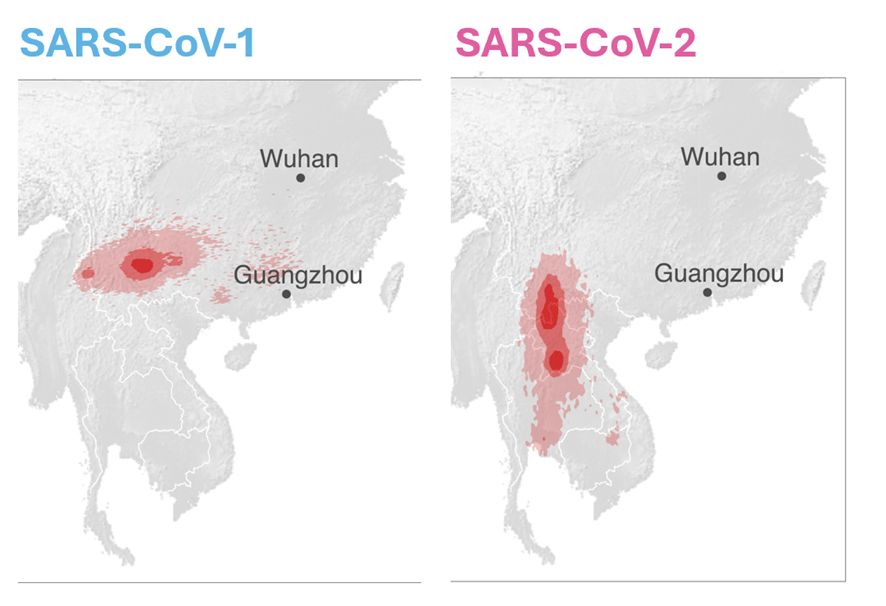
When focusing on the spike and RBD, one virus, WZ2, sampled in Zhejiang, is the single closest relative.


When focusing on the spike and RBD, one virus, WZ2, sampled in Zhejiang, is the single closest relative.
Aside from enjoying the weather and the wine I'll also be presenting some exciting (and still unpublished) work on novel sarbecovirus discovery! 🍷🏖️🦇

Aside from enjoying the weather and the wine I'll also be presenting some exciting (and still unpublished) work on novel sarbecovirus discovery! 🍷🏖️🦇
An exceptionally important topic to global health, especially in the current political climate...

An exceptionally important topic to global health, especially in the current political climate...
Unfortunately had to dip early to get to Oxford! Giving a talk on structural phylo and discussing some exciting projects with @mghafari.bsky.social tomorrow!👀👀


Unfortunately had to dip early to get to Oxford! Giving a talk on structural phylo and discussing some exciting projects with @mghafari.bsky.social tomorrow!👀👀
+ #ViBioM2025 starting tonight! Can't wait to see lots more friends! 😁😁


+ #ViBioM2025 starting tonight! Can't wait to see lots more friends! 😁😁






Looking forward to meeting lots of cool people at @recombseq.bsky.social tomorrow and chat all about AI, pLMs, structural predictions and more! 😁
I'll be presenting a poster on our recent work, link 👇

Looking forward to meeting lots of cool people at @recombseq.bsky.social tomorrow and chat all about AI, pLMs, structural predictions and more! 😁
I'll be presenting a poster on our recent work, link 👇
@cp-cellhostmicrobe.bsky.social
we summarise these findings and lay out the plausible evolutionary scenarios they may imply! 🦇🦇
doi.org/10.1016/j.ch...
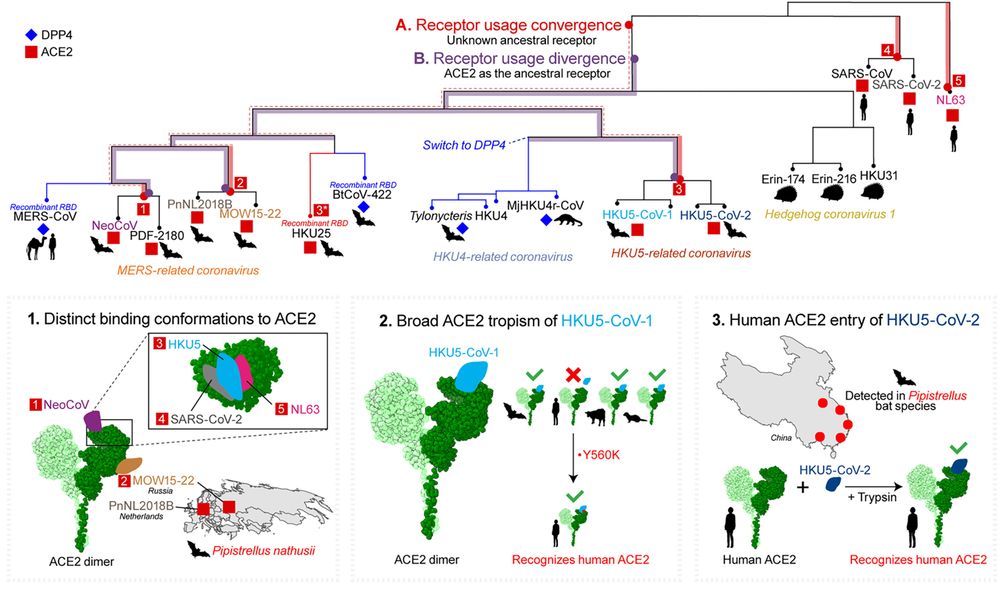
@cp-cellhostmicrobe.bsky.social
we summarise these findings and lay out the plausible evolutionary scenarios they may imply! 🦇🦇
doi.org/10.1016/j.ch...


