
Sergey Ovchinnikov
@sokrypton.org
Scientist, Assistant Professor at MIT biology, #FirstGen
Maybe something akin to paper2agent:
huggingface.co/papers/2509....
huggingface.co/papers/2509....

Paper page - Paper2Agent: Reimagining Research Papers As Interactive and Reliable AI
Agents
Join the discussion on this paper page
huggingface.co
September 11, 2025 at 4:29 PM
Maybe something akin to paper2agent:
huggingface.co/papers/2509....
huggingface.co/papers/2509....
hmmm... any ideas why mpnn would make things worse for af2, but make things about the same as af2 when used with boltz?
September 2, 2025 at 10:55 PM
hmmm... any ideas why mpnn would make things worse for af2, but make things about the same as af2 when used with boltz?
Yeah, we would expect the pseudo-likelihood to be maximized for best paired MSA!
August 6, 2025 at 12:46 AM
Yeah, we would expect the pseudo-likelihood to be maximized for best paired MSA!
We'll add an option to add custom MSA inputs to the notebook (later tonight). 😎
August 5, 2025 at 5:45 PM
We'll add an option to add custom MSA inputs to the notebook (later tonight). 😎
It was expected. The surprising part for me was that AlphaFold doesn't seem to care....
See study here from @lindseyguan.bsky.social
www.biorxiv.org/content/10.1...
This makes me wonder if the reason it doesn't seem to care is because it was trained on and evaluated on poorly paired MSAs. 🤔
See study here from @lindseyguan.bsky.social
www.biorxiv.org/content/10.1...
This makes me wonder if the reason it doesn't seem to care is because it was trained on and evaluated on poorly paired MSAs. 🤔

How AlphaFold and related models predict protein-peptide complex structures
Protein-peptide interactions mediate many biological processes, and access to accurate structural models, through experimental determination or reliable computational prediction, is essential for unde...
www.biorxiv.org
August 5, 2025 at 3:58 PM
It was expected. The surprising part for me was that AlphaFold doesn't seem to care....
See study here from @lindseyguan.bsky.social
www.biorxiv.org/content/10.1...
This makes me wonder if the reason it doesn't seem to care is because it was trained on and evaluated on poorly paired MSAs. 🤔
See study here from @lindseyguan.bsky.social
www.biorxiv.org/content/10.1...
This makes me wonder if the reason it doesn't seem to care is because it was trained on and evaluated on poorly paired MSAs. 🤔
2Y69 is technically "prokaryotic" as it's from the mitochondria. 🧐
August 5, 2025 at 1:27 PM
2Y69 is technically "prokaryotic" as it's from the mitochondria. 🧐
We find this to be true across a number of targets. Where method used to pair sequences and filter them makes a big difference. This we find to be important when trying to disentangle paralogs from orthologs (4/4).

August 5, 2025 at 7:39 AM
We find this to be true across a number of targets. Where method used to pair sequences and filter them makes a big difference. This we find to be important when trying to disentangle paralogs from orthologs (4/4).
The big difference is in the pairing. The MMseqs2 server pairs sequences based on species, while our old HHblits MSAs were paired based on genome proximity (number of genes apart). Working w/ @milot.bsky.social and @martinsteinegger.bsky.social we implemented the proximity filtering in server (3/4)

August 5, 2025 at 7:39 AM
The big difference is in the pairing. The MMseqs2 server pairs sequences based on species, while our old HHblits MSAs were paired based on genome proximity (number of genes apart). Working w/ @milot.bsky.social and @martinsteinegger.bsky.social we implemented the proximity filtering in server (3/4)
Side story: While working on the Google Colab notebook for MSA pairformer. We encountered a problem: The MMseqs2 ColabFold MSA did not show any contacts at protein interfaces, while our old HHblits alignments showed clear contacts 🫥... (2/4)
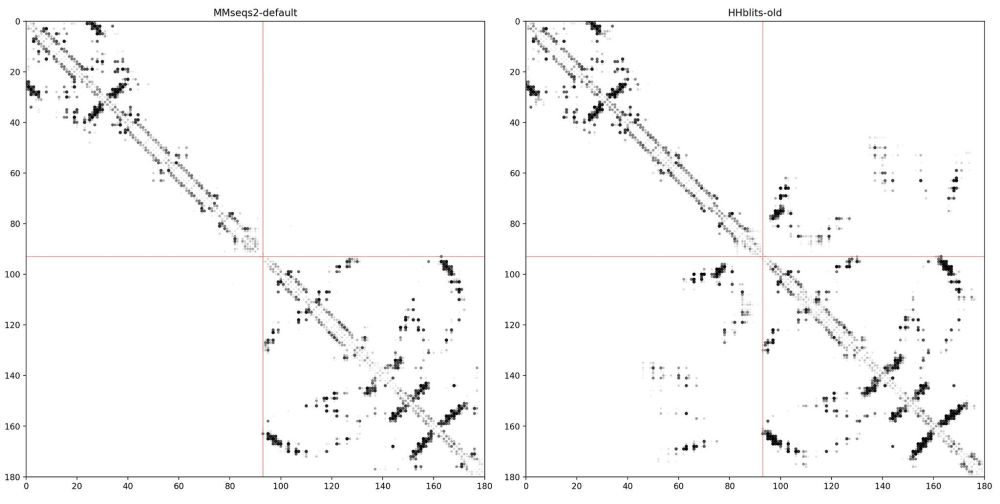
August 5, 2025 at 7:39 AM
Side story: While working on the Google Colab notebook for MSA pairformer. We encountered a problem: The MMseqs2 ColabFold MSA did not show any contacts at protein interfaces, while our old HHblits alignments showed clear contacts 🫥... (2/4)
Not sure, I was referring to the earlier observation that alphafold adjust it's confidence when additional proteins are provided. Was making a point that this is expected, and just math 🙃
August 3, 2025 at 1:27 AM
Not sure, I was referring to the earlier observation that alphafold adjust it's confidence when additional proteins are provided. Was making a point that this is expected, and just math 🙃
At bare minimum, the author should try, during inference of their own model, exclude the extra context that isn't present for gLM and see how it performs. Then I'll be convinced it's potentially learning something different.
August 2, 2025 at 8:48 PM
At bare minimum, the author should try, during inference of their own model, exclude the extra context that isn't present for gLM and see how it performs. Then I'll be convinced it's potentially learning something different.
I guess it is possible that including the full genome, the method could pick up on some key protein(s), founds across all genomes. Which would be a better classifier of phylogenetic relationships and use that for some downstream process that a smaller context of species-specific operon might miss...
August 2, 2025 at 8:46 PM
I guess it is possible that including the full genome, the method could pick up on some key protein(s), founds across all genomes. Which would be a better classifier of phylogenetic relationships and use that for some downstream process that a smaller context of species-specific operon might miss...
As it stands, it's not clear if it's actually learning anything different from gLM. For bacteria, I'm not too convinced there's context beyond +/- 15 genes for any given gene.
August 2, 2025 at 8:35 PM
As it stands, it's not clear if it's actually learning anything different from gLM. For bacteria, I'm not too convinced there's context beyond +/- 15 genes for any given gene.
the authors say "It was included in a previous version but was lost unfortunately in the editing process. " 🫠
August 2, 2025 at 8:09 PM
the authors say "It was included in a previous version but was lost unfortunately in the editing process. " 🫠

