
nature.com/articles/s41586-025-09703-7


nature.com/articles/s41586-025-09703-7
@johannavw.bsky.social presented Gemsparcl—Rapid and consistent genome clustering for navigating bacterial diversity with millions of genomes" #GI2025

@johannavw.bsky.social presented Gemsparcl—Rapid and consistent genome clustering for navigating bacterial diversity with millions of genomes" #GI2025
with myloasm"
This efficient tool uses SNPmers and leverages high quality ONT and PacBio reads for metagenome assembly.
myloasm: doi.org/10.1101/2025.09.05.674543


with myloasm"
This efficient tool uses SNPmers and leverages high quality ONT and PacBio reads for metagenome assembly.
myloasm: doi.org/10.1101/2025.09.05.674543
design choices" #GI2025 He focused more on task-specific models & showed multi-task models lack causal interpretability.
ChromBPNet: doi.org/10.1101/2024.12.25.630221
encodeproject.org


design choices" #GI2025 He focused more on task-specific models & showed multi-task models lack causal interpretability.
ChromBPNet: doi.org/10.1101/2024.12.25.630221
encodeproject.org


approaches"
Mammalian Immune Luci: doi.org/10.1093/molbev/msaf152
PatchWorkPlot: doi.org/10.1093/bioinformatics/btaf504

approaches"
Mammalian Immune Luci: doi.org/10.1093/molbev/msaf152
PatchWorkPlot: doi.org/10.1093/bioinformatics/btaf504
A very cool application of gene orthology across species for single-cell expression analysis! #orthology #GI2025 #singlecell



A very cool application of gene orthology across species for single-cell expression analysis! #orthology #GI2025 #singlecell
Chromap-QC: biorxiv.org/content/10.1101/2025.07.15.664951
Chromap: nature.com/articles/s41467-021-26865-w #GI2025


Chromap-QC: biorxiv.org/content/10.1101/2025.07.15.664951
Chromap: nature.com/articles/s41467-021-26865-w #GI2025
doi.org/10.1101/2025.05.30.656746

doi.org/10.1101/2025.05.30.656746



The first talk, by Irene Kaplow, focused on Challenges in Predicting Enhancer Activity Differences Between Species
doi.org/10.1186/s12864-022-08450-7


![OCR Ortholog Open Chromatin Status Prediction Framework Overview. a We trained a convolutional neural network (CNN) for predicting brain open chromatin using sequences underlying brain open chromatin region (OCR) orthologs in a small number of species and used the CNN to predict brain OCR ortholog open chromatin status across the species in the Zoonomia Consortium. Specifically, we used the sequences underlying the orthologs for which we have brain open chromatin data to train a CNN for predicting open chromatin. Then, we used the CNN to predict the probability of brain open chromatin for all brain OCR orthologs; predictions are illustrated on the right. Animals for which we do not have open chromatin data are in dark gray instead of black to indicate that their brain open chromatin is imputed. While we cannot evaluate the accuracy of most of our predictions, obtaining open chromatin data from most tissues in most species is infeasible, so predictions might be the best OCR annotations that we can obtain. b To demonstrate that our models can accurately predict whether sequence differences between species are associated with open chromatin differences, in addition to the evaluations described in previous work [57], we evaluated our performance on species-specific open chromatin for a species not used in model training and clade-specific open and closed chromatin for clades not used in model training. Since such regions often comprise a minority of OCR orthologs, models could obtain good overall performance while obtaining poor performance on such regions. We also evaluated our performance on tissue-specific open and closed chromatin for a tissue not used in model training, where we expect models to predict 0 if model learns sequence signatures related to the tissue used in training. c Full mouse test set and lineage-specific OCR accuracy evaluations for mouse sequence-only brain model, illustrating that, even for the best of these models,](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:jkd2l647lviezhenqed454lh/bafkreicgkhegfuippjqhdt2tkku6dr74stuf5znohmtovfk5zojt5w7xkq@jpeg)
The first talk, by Irene Kaplow, focused on Challenges in Predicting Enhancer Activity Differences Between Species
doi.org/10.1186/s12864-022-08450-7
They showed that matryoshka SAEs arxiv.org/abs/2503.17547 improves upon openSpliceAI elifesciences.org/reviewed-preprints/107454. #GI2025

They showed that matryoshka SAEs arxiv.org/abs/2503.17547 improves upon openSpliceAI elifesciences.org/reviewed-preprints/107454. #GI2025
MAPLE: nature.com/articles/s41588-023-01368-0



MAPLE: nature.com/articles/s41588-023-01368-0
A fantastic theory talk, offering intuitive insights!
Paper: doi.org/10.1101/2025.01.28.635282



A fantastic theory talk, offering intuitive insights!
Paper: doi.org/10.1101/2025.01.28.635282
doi.org/10.1371/journal.pbio.3001365 & doi.org/10.1002/cpz1.969
Software: github.com/TheBrownLab/PhyloFisher


doi.org/10.1371/journal.pbio.3001365 & doi.org/10.1002/cpz1.969
Software: github.com/TheBrownLab/PhyloFisher
Erin Molloy delivered the first talk titled "Towards scalable reconstruction of species phylogenies under the network multispecies coalescent"
TreeQMC: doi.org/10.1093/sysbio/syaf009


Erin Molloy delivered the first talk titled "Towards scalable reconstruction of species phylogenies under the network multispecies coalescent"
TreeQMC: doi.org/10.1093/sysbio/syaf009
Paper: arxiv.org/abs/2509.23426
Platform: aiscientist.tools



Paper: arxiv.org/abs/2509.23426
Platform: aiscientist.tools
based multiplex De Bruijn graphs
The goal is to perform local haplotype assembly by iteratively growing lists of k’ consecutive closed syncmers.

based multiplex De Bruijn graphs
The goal is to perform local haplotype assembly by iteratively growing lists of k’ consecutive closed syncmers.
www.biorxiv.org/content/10.1...
This work tackles the issue of Mash which ignores repeats in the genome, providing better distance estimation #GI2025


www.biorxiv.org/content/10.1...
This work tackles the issue of Mash which ignores repeats in the genome, providing better distance estimation #GI2025
They devised a bidirectional message-passing procedure in GNN for the problem of genome assembly




They devised a bidirectional message-passing procedure in GNN for the problem of genome assembly
MetaGraph is a highly compressed representation
of all public biological sequences!
nature.com/articles/s41586-025-09603-w
Try it online: metagraph.ethz.ch/search
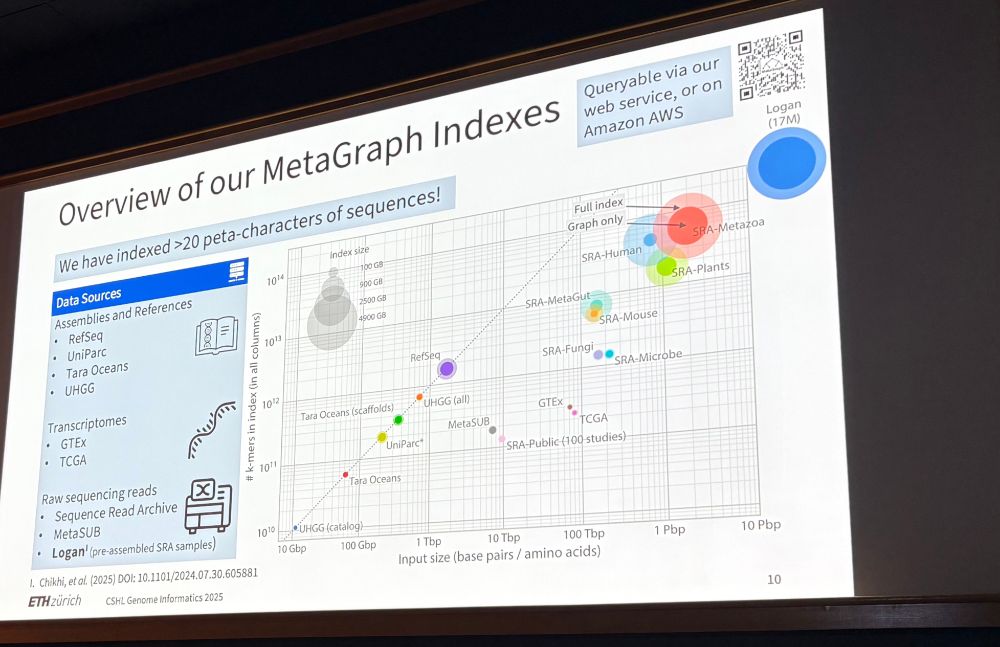


MetaGraph is a highly compressed representation
of all public biological sequences!
nature.com/articles/s41586-025-09603-w
Try it online: metagraph.ethz.ch/search
They developed a new algorithm to decompose a directed graph (generated from reads) into a minimum # weighted paths, with application to metagenome and transcriptome assembly.


They developed a new algorithm to decompose a directed graph (generated from reads) into a minimum # weighted paths, with application to metagenome and transcriptome assembly.
genome.cshlp.org/content/33/7/1175
providing theoretical guarantees for the popular seed-chain-extend


genome.cshlp.org/content/33/7/1175
providing theoretical guarantees for the popular seed-chain-extend
It uses k-mer conservation to annotate genomic variation across hundreds of genomes, followed by normalization of k-mer profiles to identify introgression events
github.com/kjenike/pana... #GI2025



It uses k-mer conservation to annotate genomic variation across hundreds of genomes, followed by normalization of k-mer profiles to identify introgression events
github.com/kjenike/pana... #GI2025

