

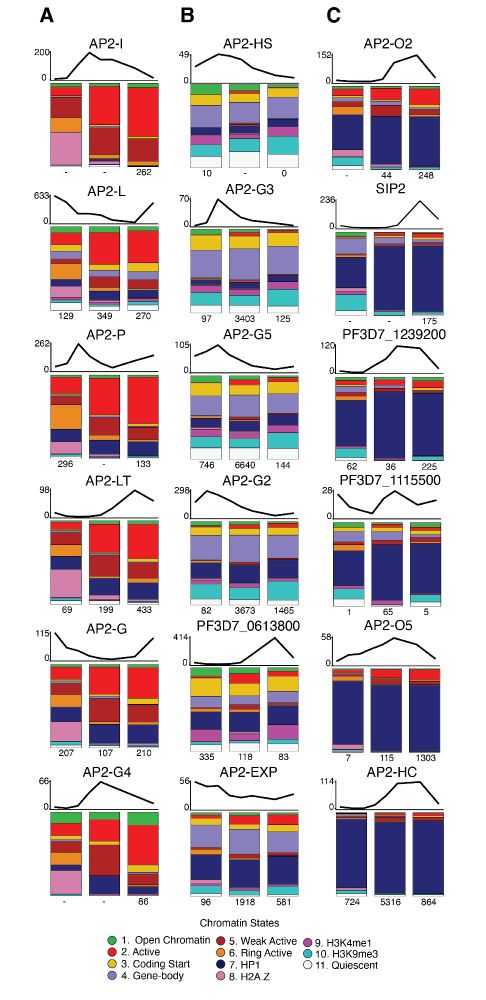


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...









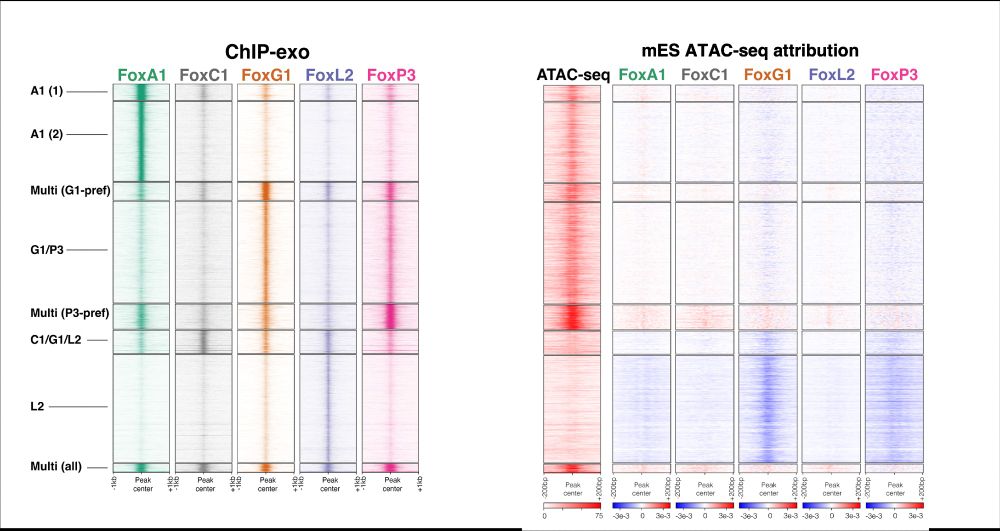






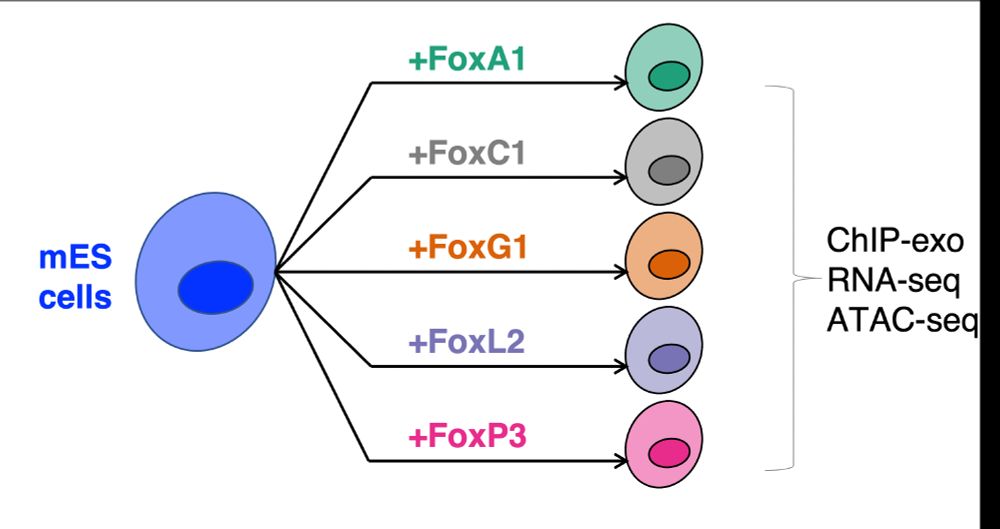

If eLife want to make the new model work, they should give better summaries of the review outcome.

If eLife want to make the new model work, they should give better summaries of the review outcome.
To turn the filters off, just select no language in settings:

To turn the filters off, just select no language in settings:

