Sean
@seanlzheng.bsky.social
Dad 👶🏻🐶🐶| Academic cardiologist ❤️ | Genomics 🧬 | Amateur allotmenteer 👨🌾
See the attached 🧵 for some of our main takeaways
🧵 Polygenic scores in hypertrophic cardiomyopathy
📎 Paper: www.nature.com/articles/s41... in @naturegenet.bsky.social
We created PGS using sumstats from our linked GWAS study www.nature.com/articles/s41..., evaluating it across a range of clinical settings in several cohorts. 1/n
📎 Paper: www.nature.com/articles/s41... in @naturegenet.bsky.social
We created PGS using sumstats from our linked GWAS study www.nature.com/articles/s41..., evaluating it across a range of clinical settings in several cohorts. 1/n
February 18, 2025 at 5:24 PM
See the attached 🧵 for some of our main takeaways
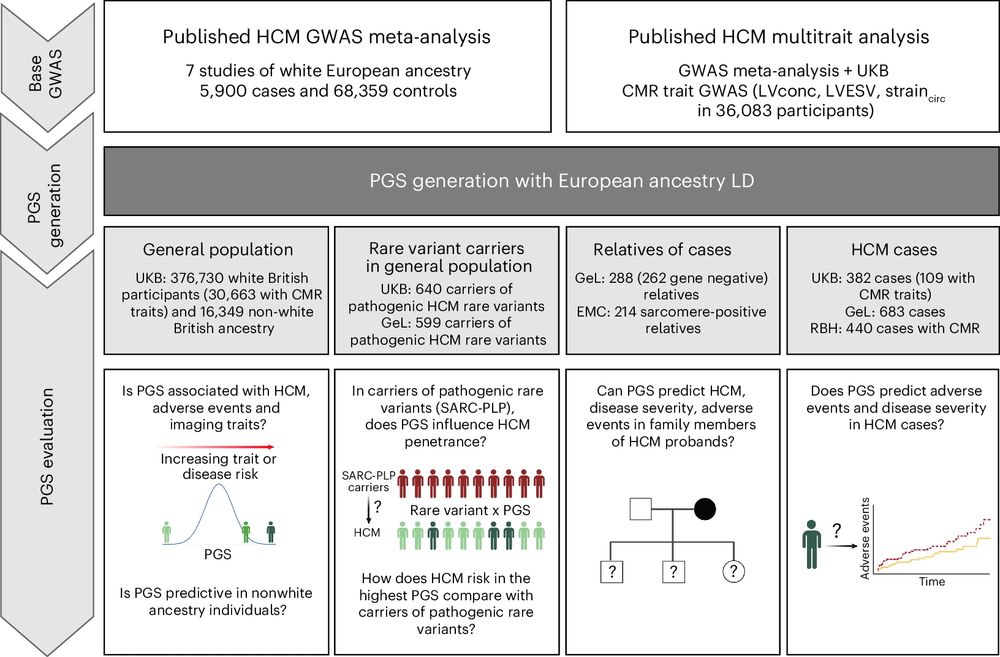
And finally, although many people with HCM lead relatively normal lives, some unfortunately develop CV complications. In >1K people with HCM in UK Biobank and 100K, PGS acts as a novel biomarker stratifying survival and MACE after diagnosis. Future work needed to fit this into clinical models. n/n
February 18, 2025 at 3:29 PM
And finally, although many people with HCM lead relatively normal lives, some unfortunately develop CV complications. In >1K people with HCM in UK Biobank and 100K, PGS acts as a novel biomarker stratifying survival and MACE after diagnosis. Future work needed to fit this into clinical models. n/n
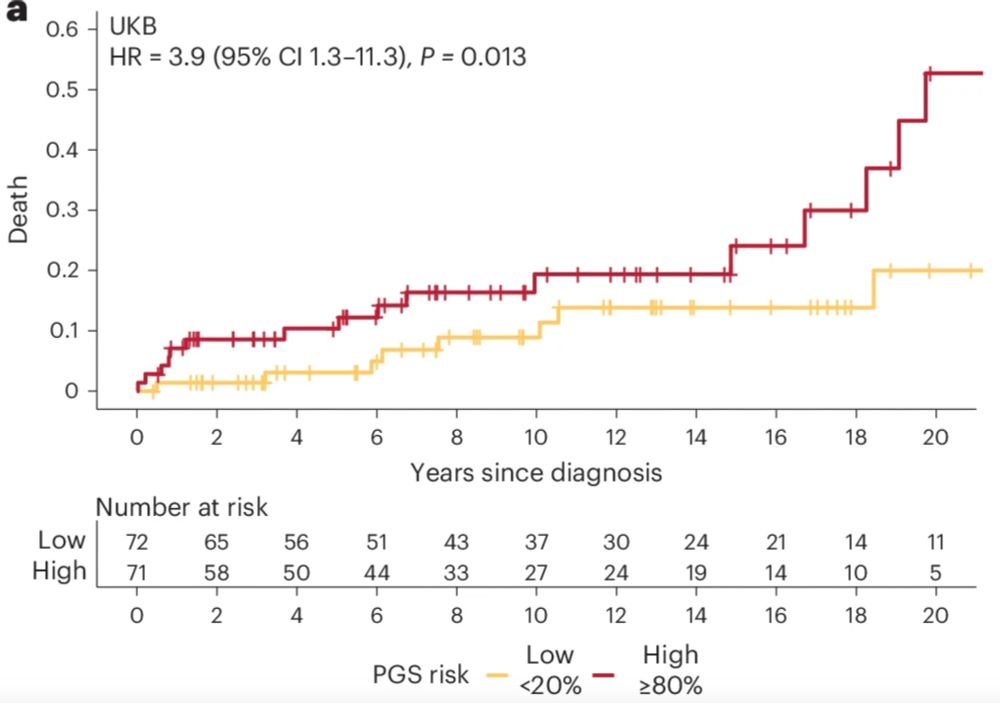
An area of clinical importance is risk assessment in patient’s families. Most relatives who are pheno-ve remain under long-term surveillance. PGS can stratify future HCM risk, HCM severity, and adverse CV outcomes among relatives, providing yet another opportunity for personalised care. 4/n
February 18, 2025 at 3:27 PM
An area of clinical importance is risk assessment in patient’s families. Most relatives who are pheno-ve remain under long-term surveillance. PGS can stratify future HCM risk, HCM severity, and adverse CV outcomes among relatives, providing yet another opportunity for personalised care. 4/n
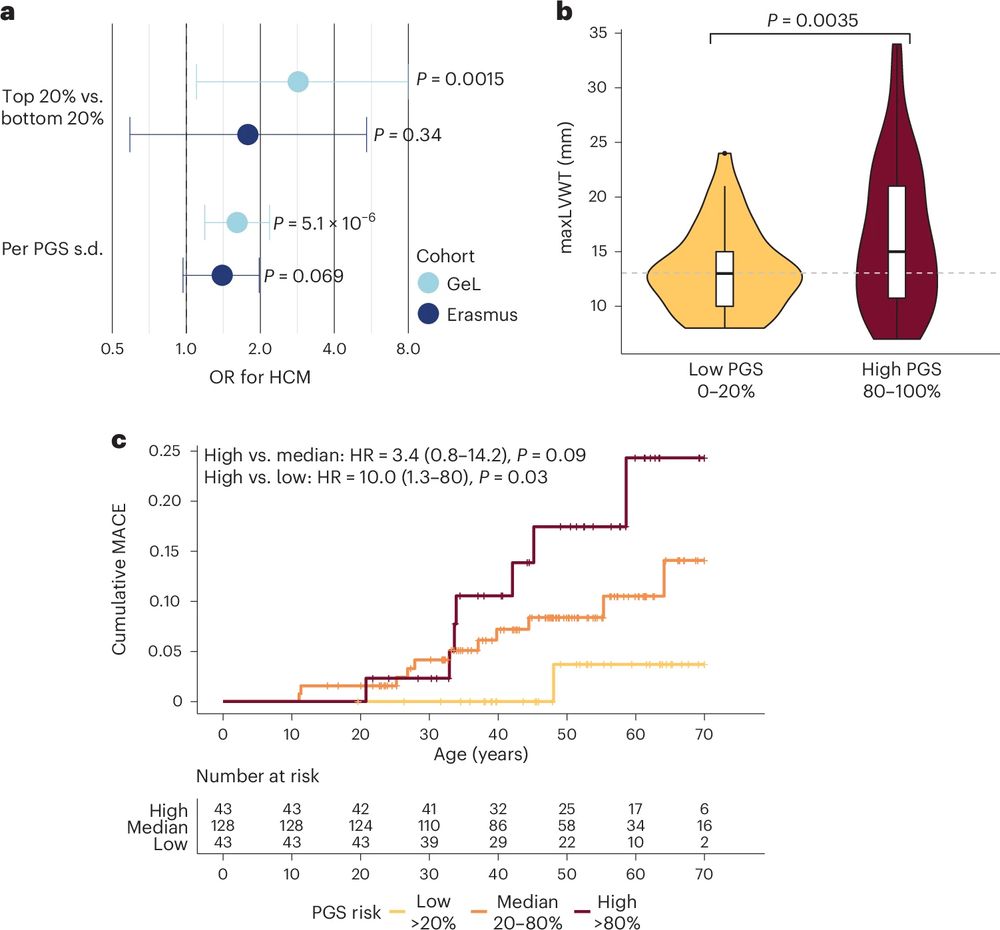
Pathogenic variant carriers in HCM genes don’t all go on to develop HCM (incompletely penetrant). In >1.2K carriers from UKB + 100K, PGS acts as a key modifier of rare variant effects = potential to guide personalised surveillance/early intervention strategies in secondary finding settings. 3/n
February 18, 2025 at 3:26 PM
Pathogenic variant carriers in HCM genes don’t all go on to develop HCM (incompletely penetrant). In >1.2K carriers from UKB + 100K, PGS acts as a key modifier of rare variant effects = potential to guide personalised surveillance/early intervention strategies in secondary finding settings. 3/n
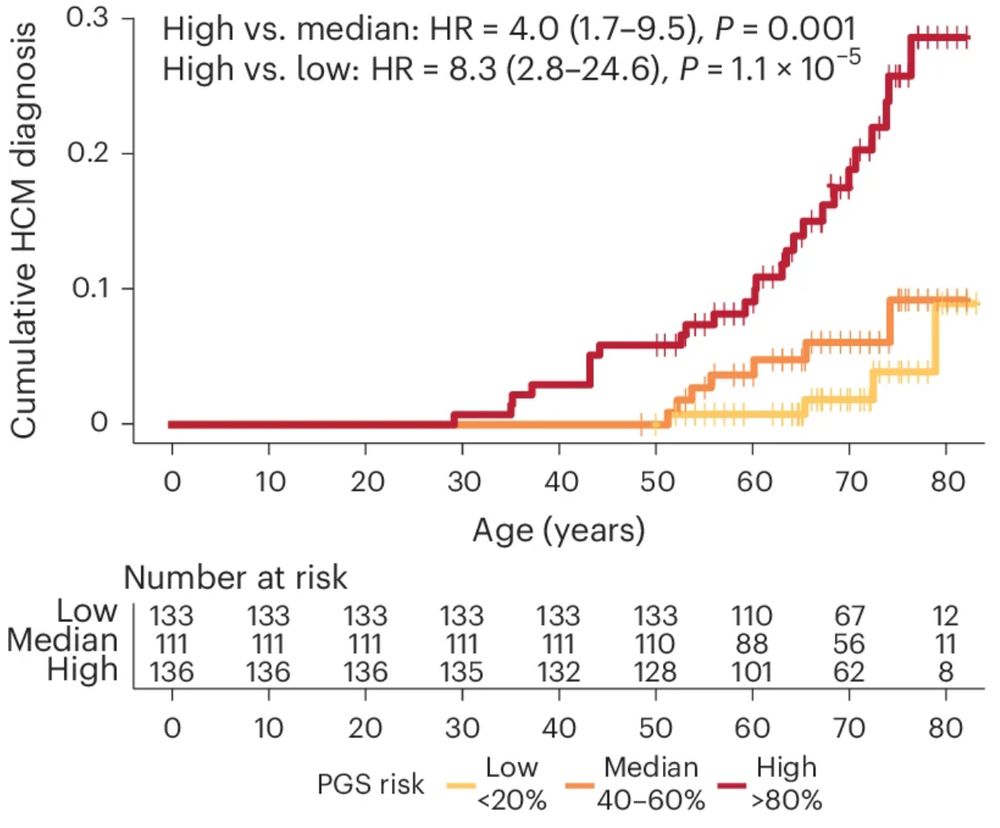
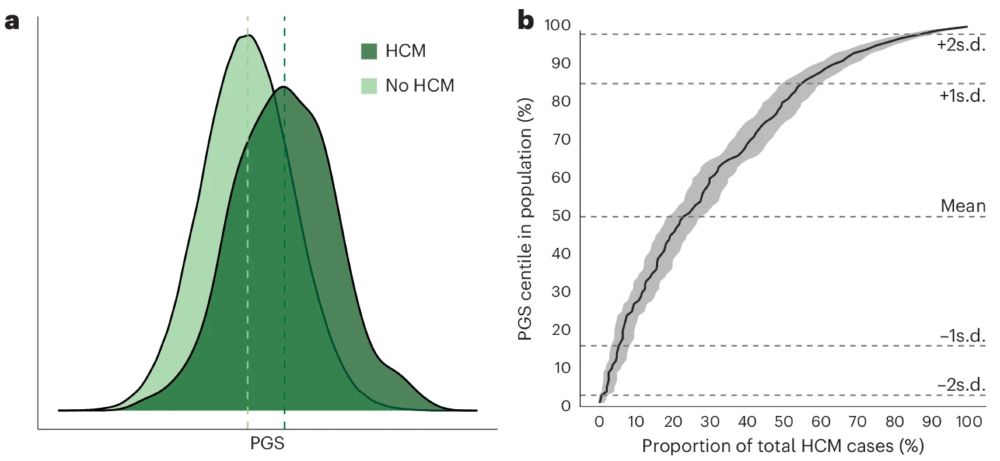
We show in the UK Biobank that PGS associates with HCM, with 15-fold increased risk of HCM for those with scores in top 1% vs. mean. Almost half of all people with HCM have a score >1SD from the mean, ~20% >2sd. 2/n
February 18, 2025 at 3:24 PM
We show in the UK Biobank that PGS associates with HCM, with 15-fold increased risk of HCM for those with scores in top 1% vs. mean. Almost half of all people with HCM have a score >1SD from the mean, ~20% >2sd. 2/n
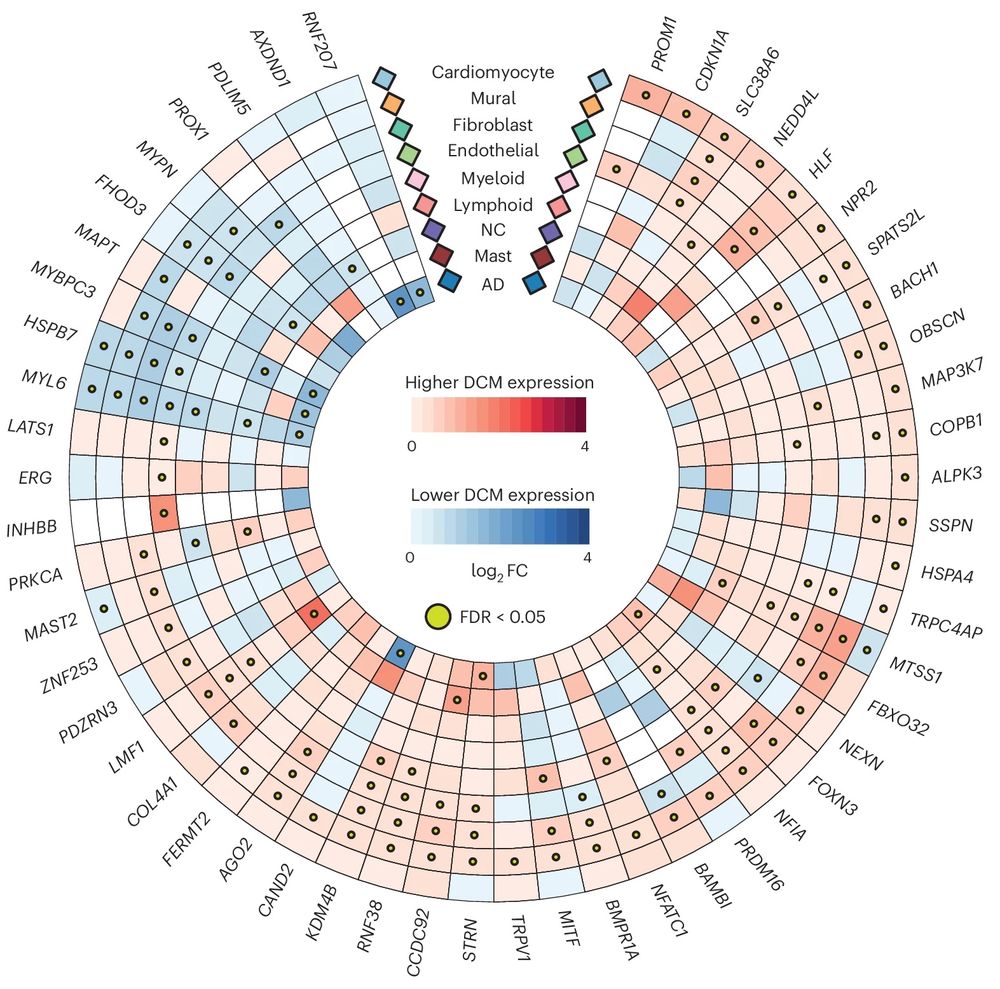
And finally, using snRNA-seq of 52 end-stage heart failure DCM samples, we identify the causal cell types, and explore changes in expression of GWAS effector genes in disease, and identify important intercellular interactions for several of the genes (COL4A1, BMPR1A, etc.)
5/5
5/5
November 21, 2024 at 1:53 PM
And finally, using snRNA-seq of 52 end-stage heart failure DCM samples, we identify the causal cell types, and explore changes in expression of GWAS effector genes in disease, and identify important intercellular interactions for several of the genes (COL4A1, BMPR1A, etc.)
5/5
5/5
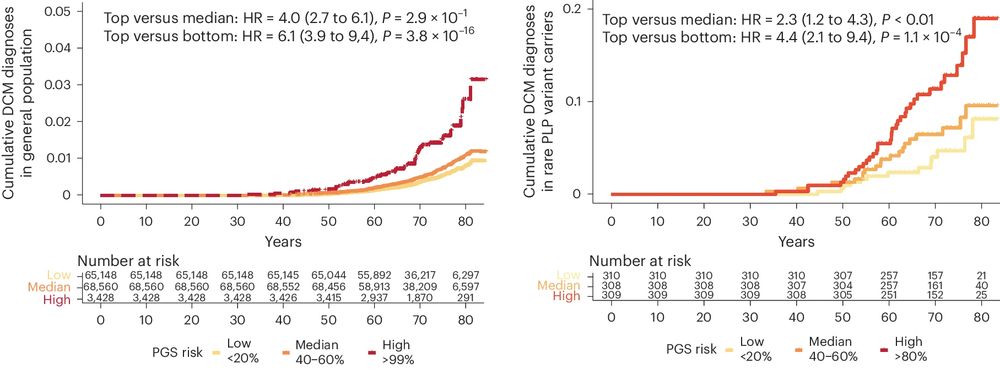
Having highlighted the importance of SNPs in DCM risk in this study, we next created a polygenic risk score and show that it predicts DCM in the population, and in 1,546 carriers of rare DCM-causing variants.
4/5
4/5
November 21, 2024 at 1:49 PM
Having highlighted the importance of SNPs in DCM risk in this study, we next created a polygenic risk score and show that it predicts DCM in the population, and in 1,546 carriers of rare DCM-causing variants.
4/5
4/5
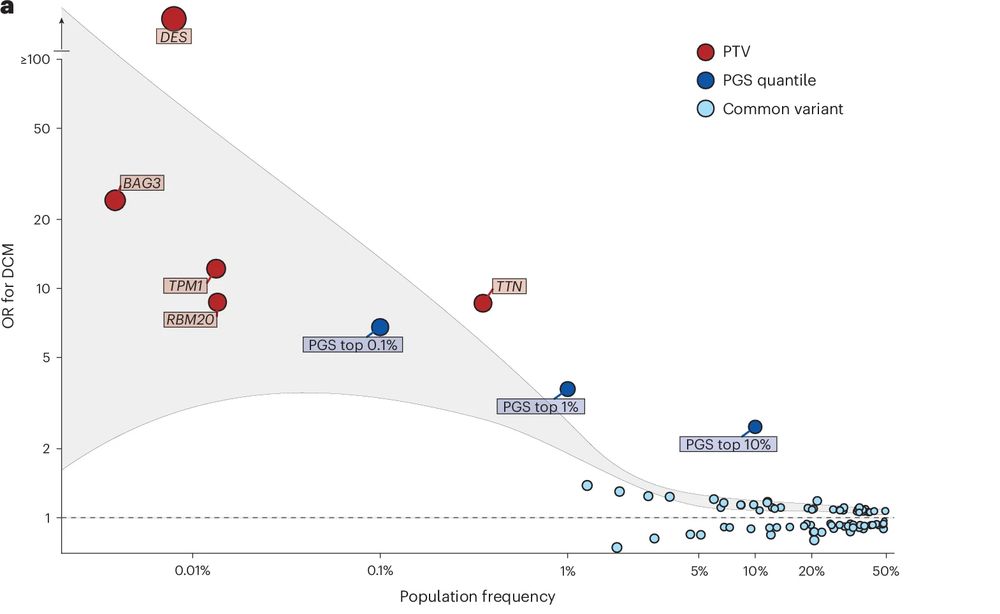
Highlighting overlap of common and rare variant causes of DCM across the spectrum of allele frequencies, 7 genes known to cause DCM were identified. Using UKBB and 100K Genomes Project we also discover 3 rare novel genetic causes of DCM (MAP3K7, SSPN, and NEDD4L).
3/5
3/5
November 21, 2024 at 1:47 PM
Highlighting overlap of common and rare variant causes of DCM across the spectrum of allele frequencies, 7 genes known to cause DCM were identified. Using UKBB and 100K Genomes Project we also discover 3 rare novel genetic causes of DCM (MAP3K7, SSPN, and NEDD4L).
3/5
3/5
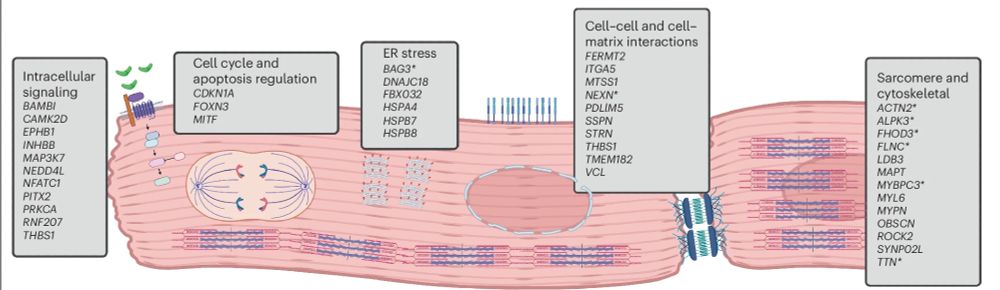
We incorporate 8 in silico tools to identify effector genes throughout the genome, and in the process highlight key biological processes involved in DCM risk - including cellular adhesion, sarcomeric function, cell signalling, and ER stress.
2/5
2/5
November 21, 2024 at 1:44 PM
We incorporate 8 in silico tools to identify effector genes throughout the genome, and in the process highlight key biological processes involved in DCM risk - including cellular adhesion, sarcomeric function, cell signalling, and ER stress.
2/5
2/5
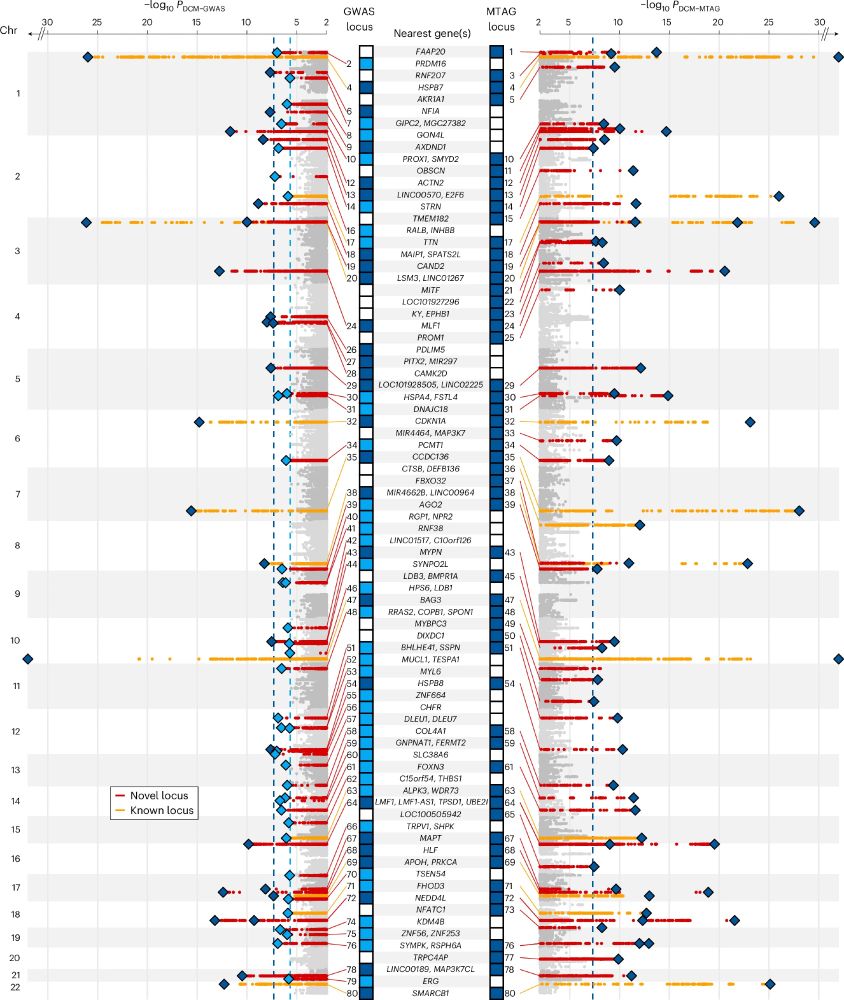
We analyse genetic data from >14K people with DCM and >1.2M controls, boosting power with multi-trait analysis incorporating CMR traits from 36K people. We find 80 risk loci (many novel) associated with DCM.
1/5
1/5
November 21, 2024 at 1:41 PM
We analyse genetic data from >14K people with DCM and >1.2M controls, boosting power with multi-trait analysis incorporating CMR traits from 36K people. We find 80 risk loci (many novel) associated with DCM.
1/5
1/5

