
Sasha Gusev
@sashagusevposts.bsky.social
Statistical geneticist. Associate Prof at Dana-Farber / Harvard Medical School.
www.gusevlab.org
www.gusevlab.org
I think about this every time I shave:

September 3, 2025 at 3:50 AM
I think about this every time I shave:
That's a great idea, and looks something like the below. I'm working on the size scaling a bit more to convey the point more clearly

August 29, 2025 at 12:37 AM
That's a great idea, and looks something like the below. I'm working on the size scaling a bit more to convey the point more clearly
So the mystery remains. It is tempting to conclude that biological epistasis is widespread but mostly gets mapped to statistical additivity. But that does not explain the deviations from additivity often observed in twins. /x

August 27, 2025 at 8:41 PM
So the mystery remains. It is tempting to conclude that biological epistasis is widespread but mostly gets mapped to statistical additivity. But that does not explain the deviations from additivity often observed in twins. /x
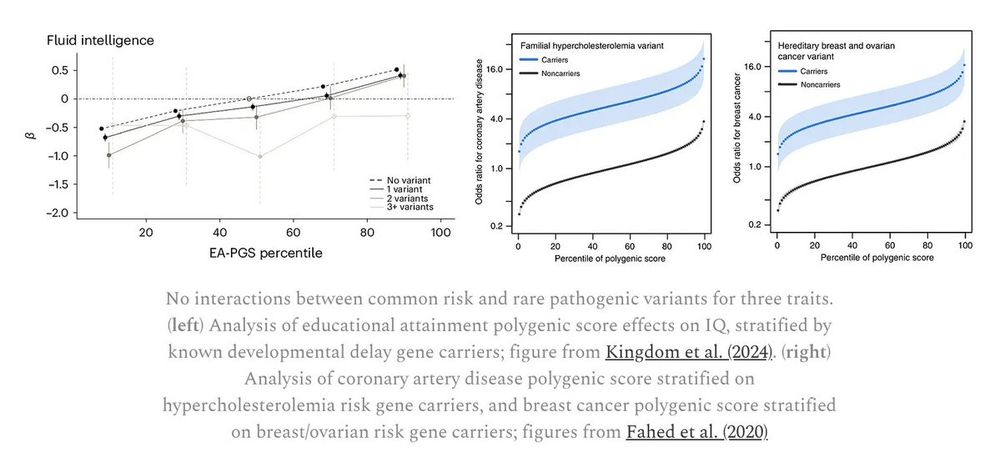
There is no statistical power to identify individual rare var interactions. But interactions between rare variants and common polygenic scores can be tested and show ... nothing. This is genuinely surprising: large deleterious effects simply add up with common polygenic burden.
August 27, 2025 at 8:41 PM
There is no statistical power to identify individual rare var interactions. But interactions between rare variants and common polygenic scores can be tested and show ... nothing. This is genuinely surprising: large deleterious effects simply add up with common polygenic burden.
So do we see non-additive epistasis in real data? For common variants, not really. Dominance has largely been ruled out, and various attempts to estimate components of pairwise epistasis have found little to none.
Could it be hiding among rare variants?
Could it be hiding among rare variants?

August 27, 2025 at 8:41 PM
So do we see non-additive epistasis in real data? For common variants, not really. Dominance has largely been ruled out, and various attempts to estimate components of pairwise epistasis have found little to none.
Could it be hiding among rare variants?
Could it be hiding among rare variants?
Finally, where epistasis really matters is in the context of interventions. If non-additive epistasis is substantial, then acting on the additive genetic mechanisms will frequently produce highly unexpected traits in individuals. Try to move a trait down but it actually goes up:

August 27, 2025 at 8:41 PM
Finally, where epistasis really matters is in the context of interventions. If non-additive epistasis is substantial, then acting on the additive genetic mechanisms will frequently produce highly unexpected traits in individuals. Try to move a trait down but it actually goes up:
What about natural selection? For the short-term response, all that matters is additive/narrow-sense heritability. But the long-term response can be very different depending on whether epistasis is aligned with or against fitness:

August 27, 2025 at 8:41 PM
What about natural selection? For the short-term response, all that matters is additive/narrow-sense heritability. But the long-term response can be very different depending on whether epistasis is aligned with or against fitness:
That's at the variant level, but Zuk et al. 2012 proposed a model where a trait is defined by the lowest value of multiple heritable pathways ("each unhappy family is unhappy in its own way"). This *induces* epistasis and can similarly severely distort heritability estimates.

August 27, 2025 at 8:41 PM
That's at the variant level, but Zuk et al. 2012 proposed a model where a trait is defined by the lowest value of multiple heritable pathways ("each unhappy family is unhappy in its own way"). This *induces* epistasis and can similarly severely distort heritability estimates.
Less appreciated is the fact that this inflation has to go ~somewhere~, so it also leads to a corresponding deflation in the estimate of the shared environment. The same twin correlations can arise if [A]dditivity=74% and [C]ommon-Env=12% or from [A]=20%, [C]=30%, [D]=35%.

August 27, 2025 at 8:41 PM
Less appreciated is the fact that this inflation has to go ~somewhere~, so it also leads to a corresponding deflation in the estimate of the shared environment. The same twin correlations can arise if [A]dditivity=74% and [C]ommon-Env=12% or from [A]=20%, [C]=30%, [D]=35%.
pistasis decays very quickly with genotypic relatedness, which creates problems for family based heritability estimators. Narrow-sense heritability estimates from unrelated individuals pick up ~only the additive component, whereas estimates from twins can be severely inflated.

August 27, 2025 at 8:41 PM
pistasis decays very quickly with genotypic relatedness, which creates problems for family based heritability estimators. Narrow-sense heritability estimates from unrelated individuals pick up ~only the additive component, whereas estimates from twins can be severely inflated.
The same phenomena holds for genome-wide epistasis between many loci. As the causal variants become more common, the epistasis (A*B) can be better approximated with additivity (A+B). For uniformly selected frequencies, >80% of biological epistasis looks like additivity!

August 27, 2025 at 8:41 PM
The same phenomena holds for genome-wide epistasis between many loci. As the causal variants become more common, the epistasis (A*B) can be better approximated with additivity (A+B). For uniformly selected frequencies, >80% of biological epistasis looks like additivity!
For two loci in a simple additive model (A+B), the effects on the trait are "parallel" (left). Interactions between the loci (AxB) draw the effects away from parallel, with larger deviations for rare variants. Surprisingly, high frequency AxB looks very additive *statistically*.

August 27, 2025 at 8:41 PM
For two loci in a simple additive model (A+B), the effects on the trait are "parallel" (left). Interactions between the loci (AxB) draw the effects away from parallel, with larger deviations for rare variants. Surprisingly, high frequency AxB looks very additive *statistically*.
One of the largest twin studies of personality anticipated massive non-additive dominance/epistasis components. These have not materialized with modern data for *any trait*, no one knows why, and the entire set of findings has apparently been memory-holed.


August 17, 2025 at 6:34 PM
One of the largest twin studies of personality anticipated massive non-additive dominance/epistasis components. These have not materialized with modern data for *any trait*, no one knows why, and the entire set of findings has apparently been memory-holed.
Decrease in phenotype by intervening on additive or epistatic mechanisms in the presence of low (left) or high (right) epistasis. Red lines indicate an increase in the phenotype when a decrease was intended.

August 12, 2025 at 9:25 PM
Decrease in phenotype by intervening on additive or epistatic mechanisms in the presence of low (left) or high (right) epistasis. Red lines indicate an increase in the phenotype when a decrease was intended.
I bring this up as more embryo selection companies are advertising large relative risk reductions for traits that clearly lie on a spectrum. These reductions are in turn interpreted as probabilistic "cures" even by a fairly informed audience (e.g. astralcodexten.com/p/suddenly-t...). /x


August 10, 2025 at 7:08 PM
I bring this up as more embryo selection companies are advertising large relative risk reductions for traits that clearly lie on a spectrum. These reductions are in turn interpreted as probabilistic "cures" even by a fairly informed audience (e.g. astralcodexten.com/p/suddenly-t...). /x
When we look at the expected additional "disease-free time" for a typical person (including those who would never develop the disease in their lifetime) the risk reduction adds just a few months. In contrast, classical relative risk estimates are a sizable ~20%.

August 10, 2025 at 7:08 PM
When we look at the expected additional "disease-free time" for a typical person (including those who would never develop the disease in their lifetime) the risk reduction adds just a few months. In contrast, classical relative risk estimates are a sizable ~20%.
We can again use simulations to see what happens when risk is reduced. For a trait that mimics breast cancer incidence and a 0.5SD risk reduction, individuals who would have developed the condition instead develop it 3-6 years later in life.

August 10, 2025 at 7:08 PM
We can again use simulations to see what happens when risk is reduced. For a trait that mimics breast cancer incidence and a 0.5SD risk reduction, individuals who would have developed the condition instead develop it 3-6 years later in life.
What about diseases like cancer? A second model we can think about is a `hazard` or survival model, where individuals develop the disease at different rates. Here embryo screening essentially extends the age of onset for the disease in the offspring, rather than curing it.

August 10, 2025 at 7:08 PM
What about diseases like cancer? A second model we can think about is a `hazard` or survival model, where individuals develop the disease at different rates. Here embryo screening essentially extends the age of onset for the disease in the offspring, rather than curing it.
In the case of obesity, there's nothing magical about crossing the threshold. Such ad hoc thresholds -- sometimes called "dichotomania" -- are surprisingly common in medicine. In fact, many traits currently being screened for have clinically important sub-threshold analogs:

August 10, 2025 at 7:08 PM
In the case of obesity, there's nothing magical about crossing the threshold. Such ad hoc thresholds -- sometimes called "dichotomania" -- are surprisingly common in medicine. In fact, many traits currently being screened for have clinically important sub-threshold analogs:
In the setting of embryo selection, small changes in the underlying liability can lead to large apparent "relative risk reductions", because individuals are shifted from just above to just below the threshold. See this simulated example with Class III obesity:

August 10, 2025 at 7:08 PM
In the setting of embryo selection, small changes in the underlying liability can lead to large apparent "relative risk reductions", because individuals are shifted from just above to just below the threshold. See this simulated example with Class III obesity:
I argue that the main distinction here is the use of a statistical model that messes with our intuitions about disease. The `liability threshold model` assumes all individuals lie along a liability continuum; those above a threshold are sick and those below are perfectly healthy.

August 10, 2025 at 7:08 PM
I argue that the main distinction here is the use of a statistical model that messes with our intuitions about disease. The `liability threshold model` assumes all individuals lie along a liability continuum; those above a threshold are sick and those below are perfectly healthy.
The core issue is captured by two seminal papers. First, Karavani et al. 2019 (pubmed.ncbi.nlm.nih.gov/31761530/) found that screening embryos for continuous traits has limited utility. Then, in 2021 many of the same authors found that selection CAN reduce risk for certain diseases.


August 10, 2025 at 7:08 PM
The core issue is captured by two seminal papers. First, Karavani et al. 2019 (pubmed.ncbi.nlm.nih.gov/31761530/) found that screening embryos for continuous traits has limited utility. Then, in 2021 many of the same authors found that selection CAN reduce risk for certain diseases.
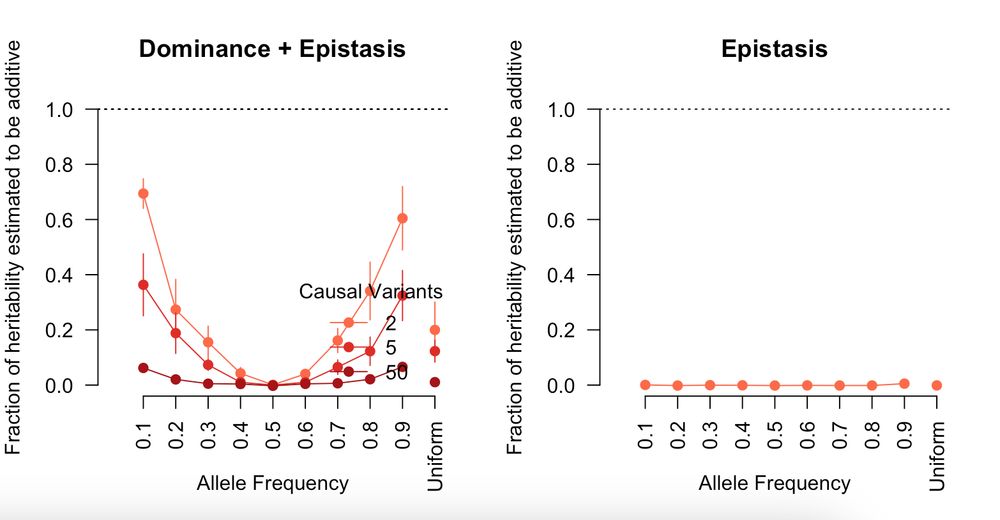
If you center the genotypes then you are effectively adding only non-additive epistasis so the additive component misses all of it (except for dominance).
August 2, 2025 at 7:47 PM
If you center the genotypes then you are effectively adding only non-additive epistasis so the additive component misses all of it (except for dominance).
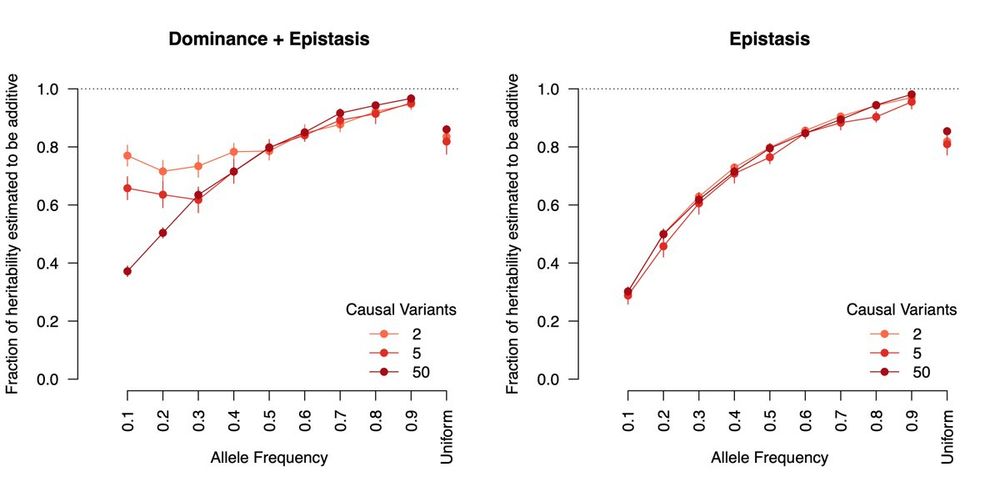
The proportion of epistatic heritability that is estimated as additive by quantitative genetic models. Epistasis deviates more from additivity for lower frequency causal alleles, but on average >80% of biological GxG will just look like statistical G.
August 2, 2025 at 3:54 PM
The proportion of epistatic heritability that is estimated as additive by quantitative genetic models. Epistasis deviates more from additivity for lower frequency causal alleles, but on average >80% of biological GxG will just look like statistical G.
A fourth employee of Heliospect is an active quantitative racism researcher, publishing with the usual race science crew on bogus Jewish polygenic score comparisons (work that was immediately and summarily rebutted: trejo.scholar.princeton.edu/sites/g/file...).



August 2, 2025 at 2:38 PM
A fourth employee of Heliospect is an active quantitative racism researcher, publishing with the usual race science crew on bogus Jewish polygenic score comparisons (work that was immediately and summarily rebutted: trejo.scholar.princeton.edu/sites/g/file...).

