https://samuel-marsh.science
17/n
17/n
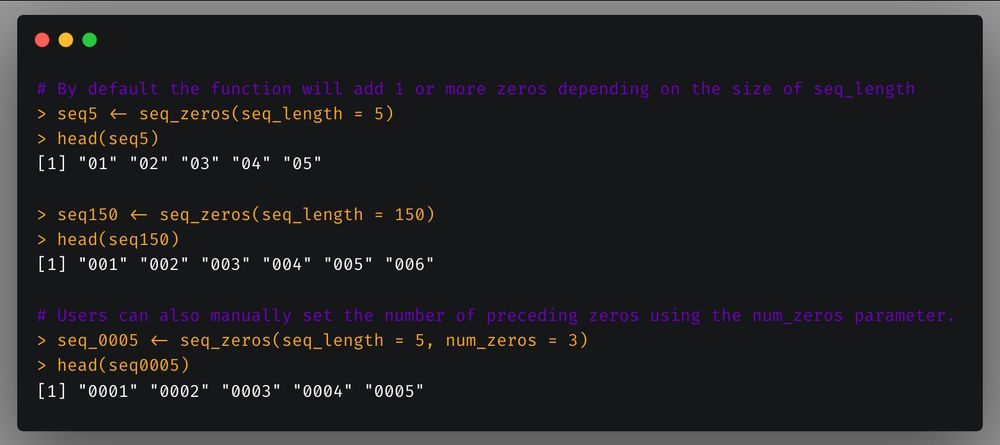
First, `Split_Vector` which allows you to split a vector in a specified number of chunks or chunks of specific length.
15/n
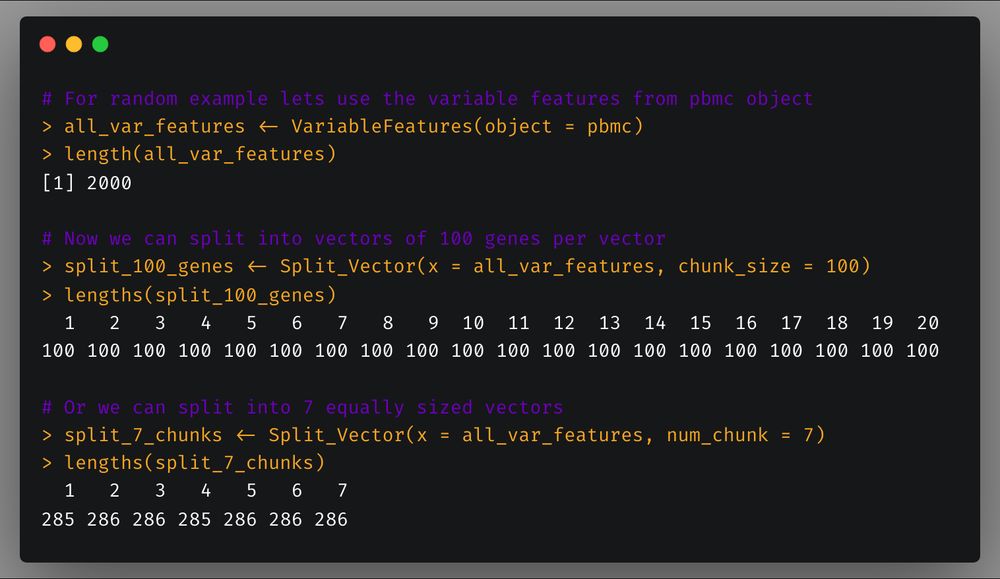
First, `Split_Vector` which allows you to split a vector in a specified number of chunks or chunks of specific length.
15/n
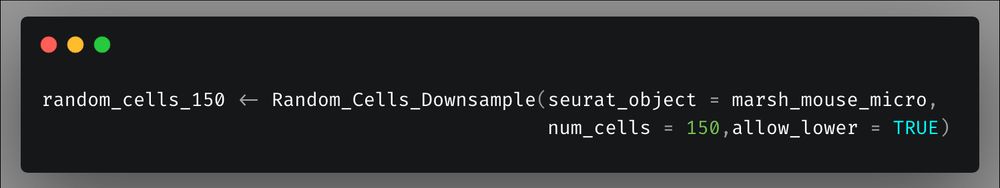
To make it even easier you can also just call `Random_Cells_Downsample` from within the `DoHeatmap` call.
14/n
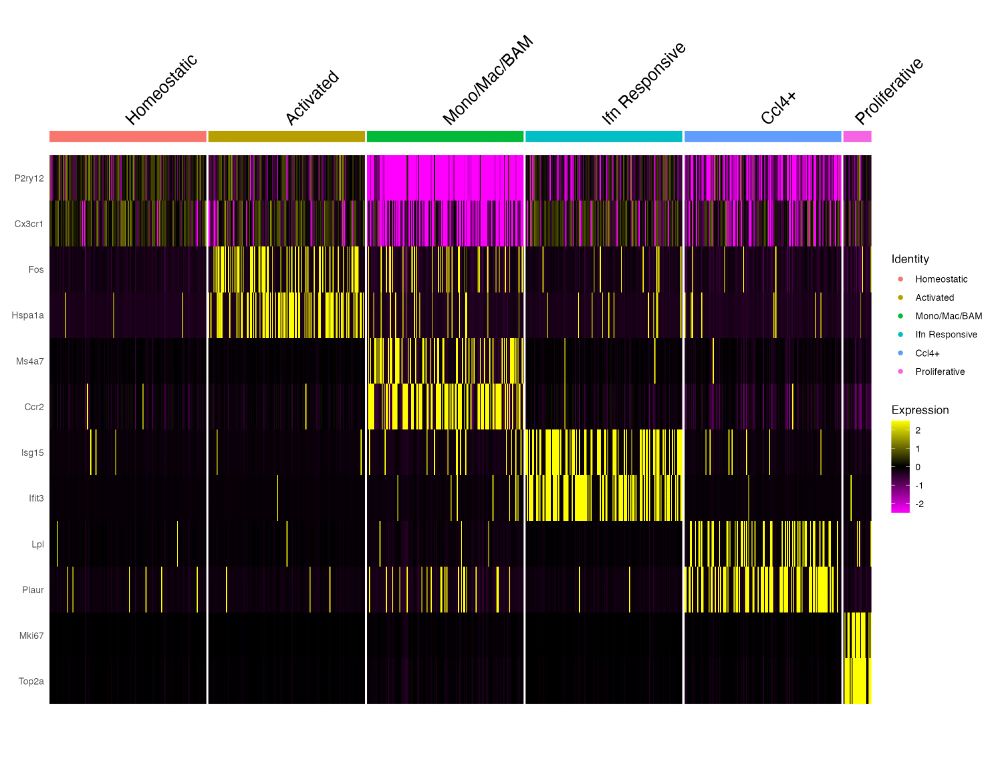
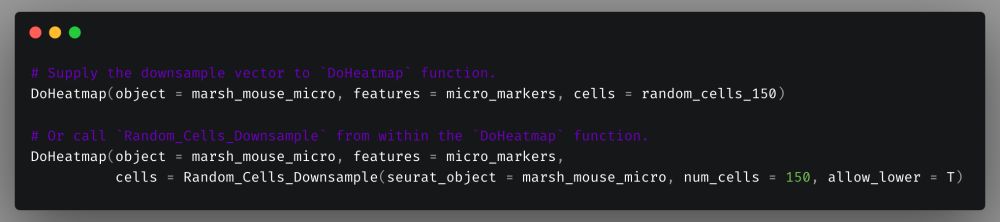
To make it even easier you can also just call `Random_Cells_Downsample` from within the `DoHeatmap` call.
14/n
13/n
13/n
12/n
12/n
9/n
9/n
`Add_Cell_QC_Metrics` now includes addition of % of hemoglobin counts as metric & chicken was added to default species.
Default species are now human, mouse, rat, drosophila, zebrafish, macaque, marmoset, & chicken.
One line will add all QC metrics to your object.
2/n
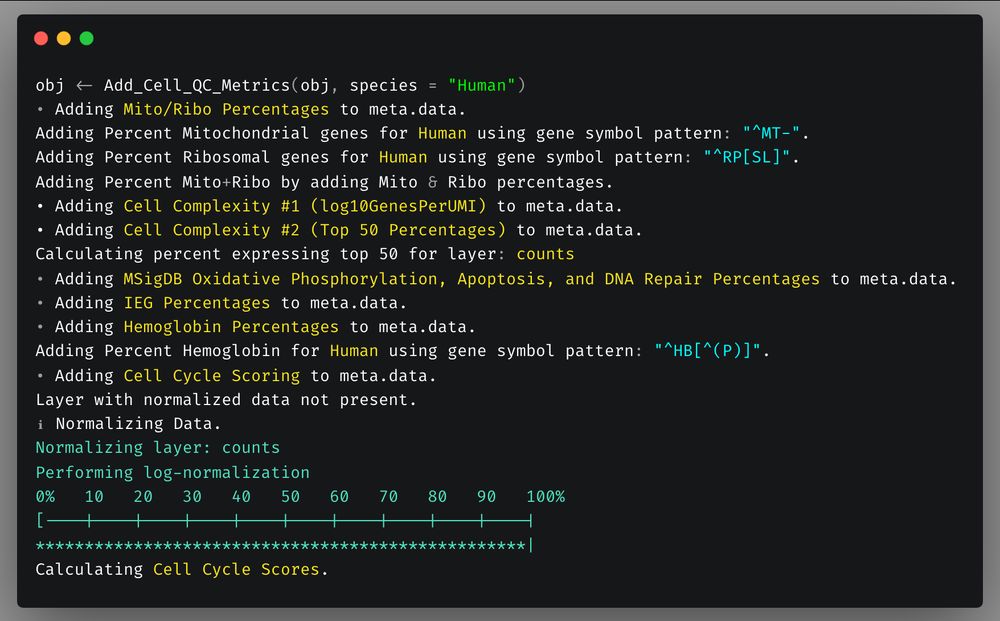
`Add_Cell_QC_Metrics` now includes addition of % of hemoglobin counts as metric & chicken was added to default species.
Default species are now human, mouse, rat, drosophila, zebrafish, macaque, marmoset, & chicken.
One line will add all QC metrics to your object.
2/n
- `QC_Histogram` to plot QC features (or any other meta data or feature/gene)
- `FeatureScatter_scCustom` joins the scCustomize family of customized Seurat plots
6/n

- `QC_Histogram` to plot QC features (or any other meta data or feature/gene)
- `FeatureScatter_scCustom` joins the scCustomize family of customized Seurat plots
6/n
- Adding boxplots to any violin plot function with `plot_boxplot`
- Just add median with `plot_median`
- Removing plot axes in any DimPlot-based function with `figure_plot`
5/n



- Adding boxplots to any violin plot function with `plot_boxplot`
- Just add median with `plot_median`
- Removing plot axes in any DimPlot-based function with `figure_plot`
5/n
v2.0.0 also introduces a one-liner to add all qc metrics in one fell swoop:
4/n

v2.0.0 also introduces a one-liner to add all qc metrics in one fell swoop:
4/n

