PhD @ MIT Computational and Systems Biology | MBA Fellow at MIT Sloan. @SabetiLab member
arxiv.org/abs/2503.16351

arxiv.org/abs/2503.16351
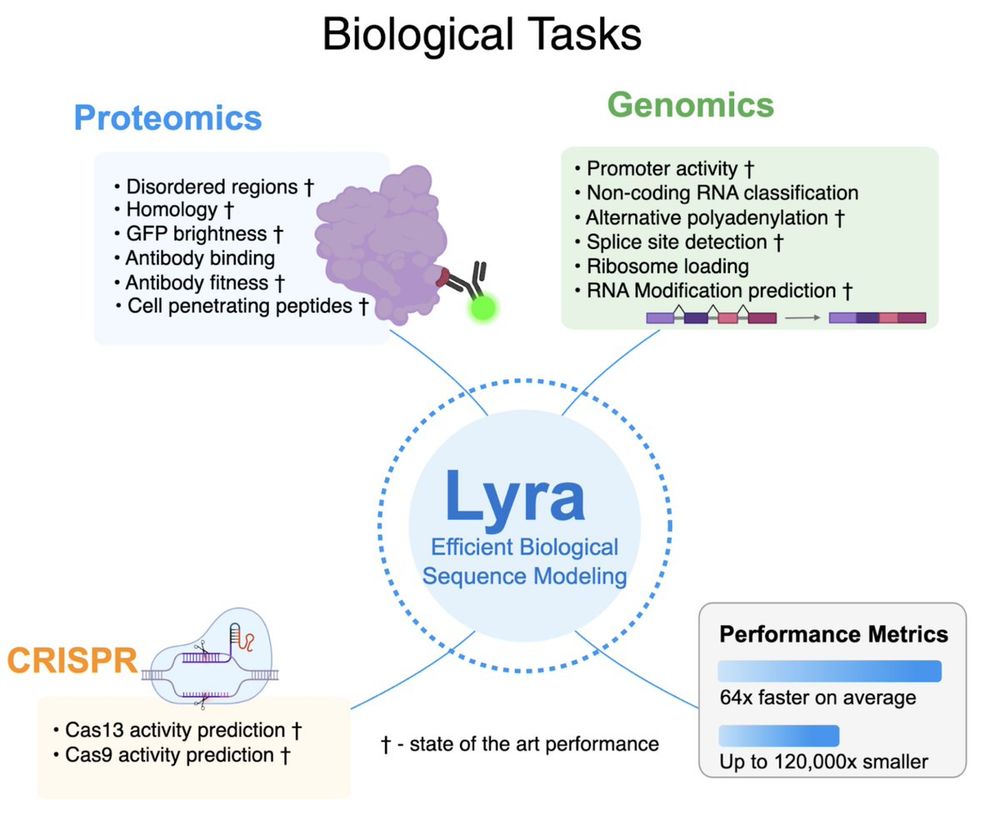
1. Proteomics
2. Genomics
3. CRISPR guide efficacy
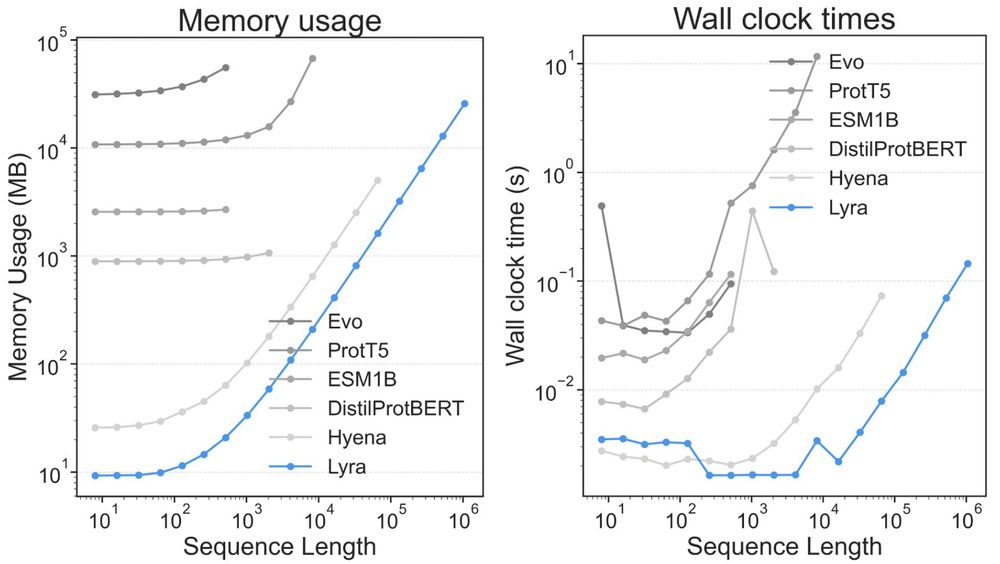
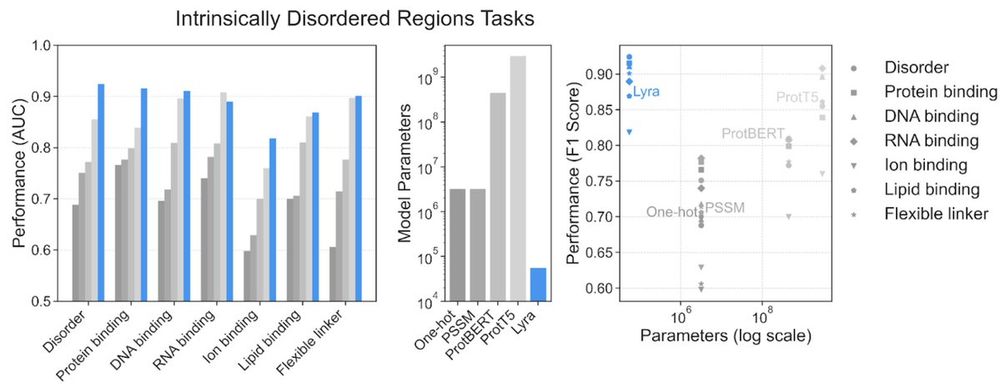
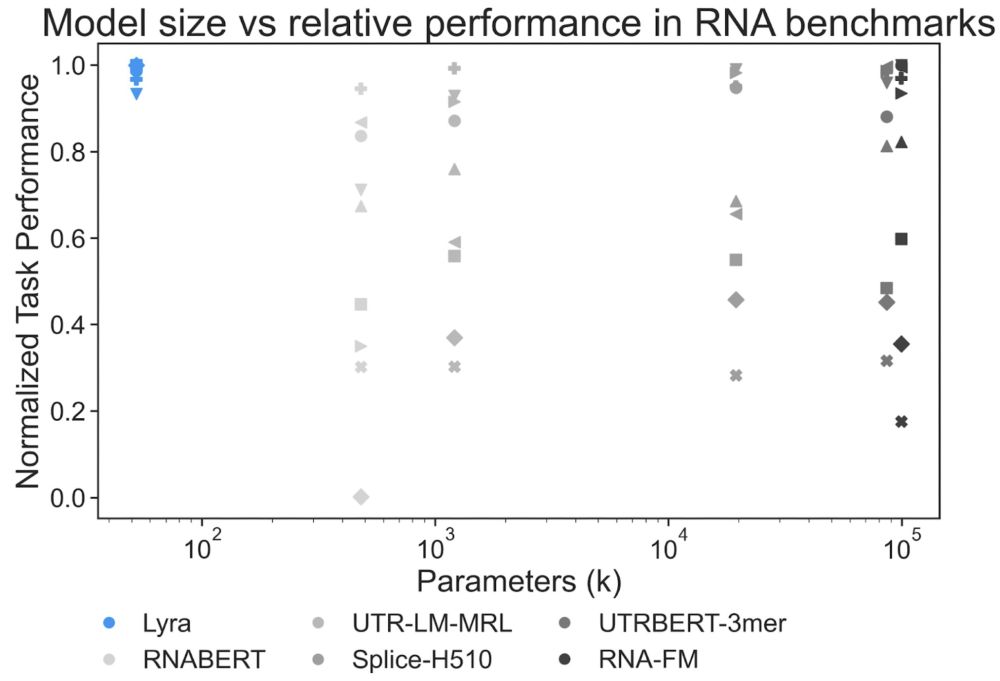
Lyra set new performance records in 79 out of 101 tasks, w/ substantially smaller models than competing architectures.
1. Proteomics
2. Genomics
3. CRISPR guide efficacy
Lyra set new performance records in 79 out of 101 tasks, w/ substantially smaller models than competing architectures.