
Samantha Graham
@samanthagraham.bsky.social
Postdoc at Brigham and Women's Hospital studying COPD | she/her | spgraham1.github.io
Thank you! Yes, we're planning on continuing to process new data on SRA and update the resource
January 23, 2025 at 4:04 PM
Thank you! Yes, we're planning on continuing to process new data on SRA and update the resource
Thanks to our many collaborators who made this possible! This has been a huge effort from our lab and others, and we couldn’t have done it without all who helped.
January 22, 2025 at 5:31 PM
Thanks to our many collaborators who made this possible! This has been a huge effort from our lab and others, and we couldn’t have done it without all who helped.
We hope this is a useful resource for the microbiome community. Links below!
Paper: www.cell.com/cell/fulltex...
Dataset: doi.org/10.5281/zeno...
Website: microbiomap.org
Code: github.com/blekhmanlab/...
Paper: www.cell.com/cell/fulltex...
Dataset: doi.org/10.5281/zeno...
Website: microbiomap.org
Code: github.com/blekhmanlab/...
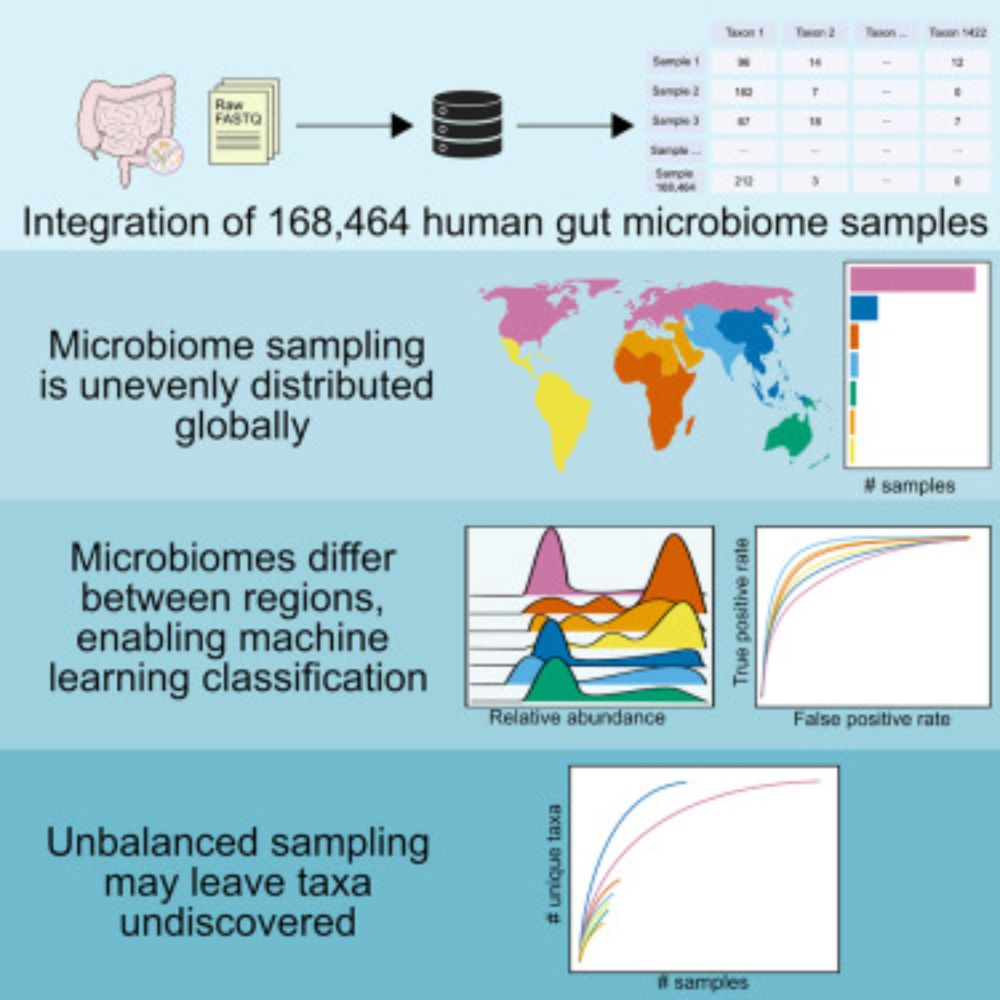
Integration of 168,000 samples reveals global patterns of the human gut microbiome
The Human Microbiome Compendium is a unified database of publicly available human
gut microbiome 16S samples, built with the integrated data from hundreds of independent
projects. The compendium is us...
www.cell.com
January 22, 2025 at 5:31 PM
We hope this is a useful resource for the microbiome community. Links below!
Paper: www.cell.com/cell/fulltex...
Dataset: doi.org/10.5281/zeno...
Website: microbiomap.org
Code: github.com/blekhmanlab/...
Paper: www.cell.com/cell/fulltex...
Dataset: doi.org/10.5281/zeno...
Website: microbiomap.org
Code: github.com/blekhmanlab/...
Altogether, this dataset shows us how microbiomes differ throughout the world, and emphasizes the importance of more even sampling moving forward. We’re excited to continue learning from the compendium, and we plan to continue to process more samples to keep it up to date with the available data.
January 22, 2025 at 5:31 PM
Altogether, this dataset shows us how microbiomes differ throughout the world, and emphasizes the importance of more even sampling moving forward. We’re excited to continue learning from the compendium, and we plan to continue to process more samples to keep it up to date with the available data.
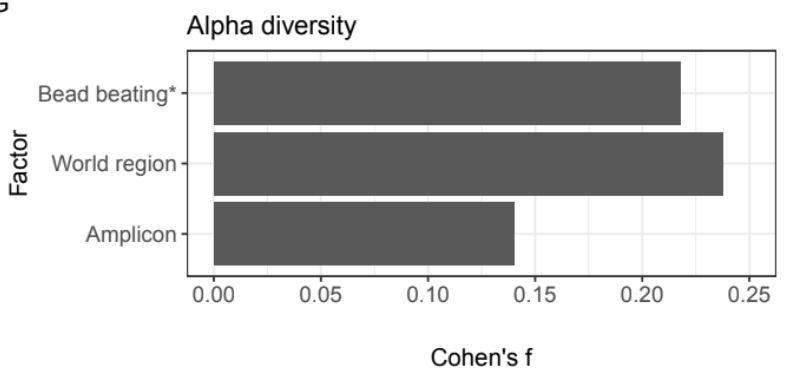
The method of DNA extraction also has a strong effect on microbiome composition. But we found that a sample’s geographic location has a bigger effect on its alpha diversity than amplicon choice or the use of bead beating, a common technique in DNA extraction.
January 22, 2025 at 5:31 PM
The method of DNA extraction also has a strong effect on microbiome composition. But we found that a sample’s geographic location has a bigger effect on its alpha diversity than amplicon choice or the use of bead beating, a common technique in DNA extraction.
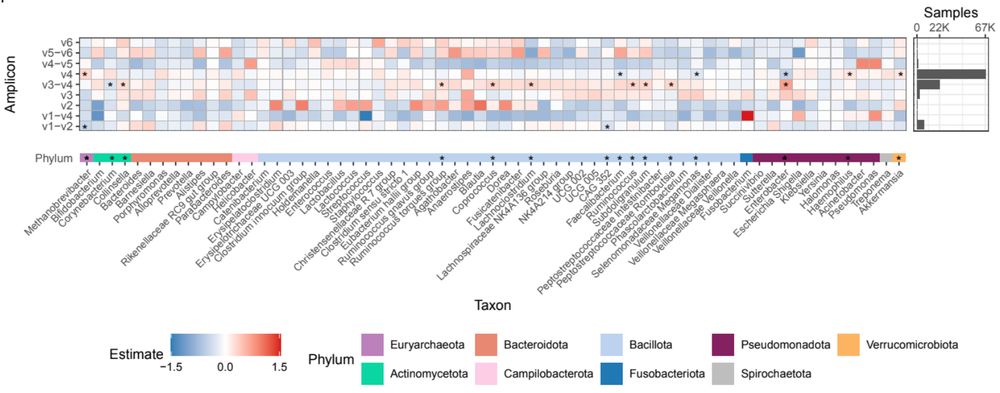
In addition to biological differences between samples, there are technical factors that can affect microbiome composition. Does the region of the 16S gene that was sequenced affect our observed results? Yes! We found genera that were differentially abundant between amplicons.
January 22, 2025 at 5:31 PM
In addition to biological differences between samples, there are technical factors that can affect microbiome composition. Does the region of the 16S gene that was sequenced affect our observed results? Yes! We found genera that were differentially abundant between amplicons.
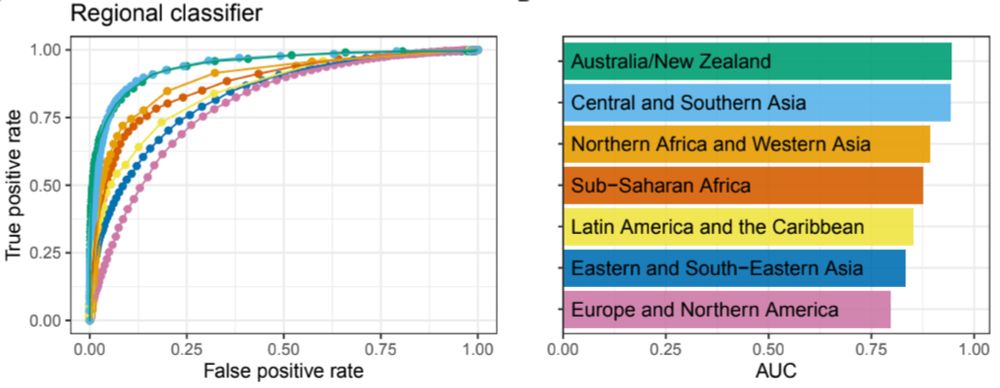
And because microbiomes from these regions are so different, we were able to train one-vs-all random forest classifiers to successfully predict where a microbiome sample came from.
January 22, 2025 at 5:31 PM
And because microbiomes from these regions are so different, we were able to train one-vs-all random forest classifiers to successfully predict where a microbiome sample came from.
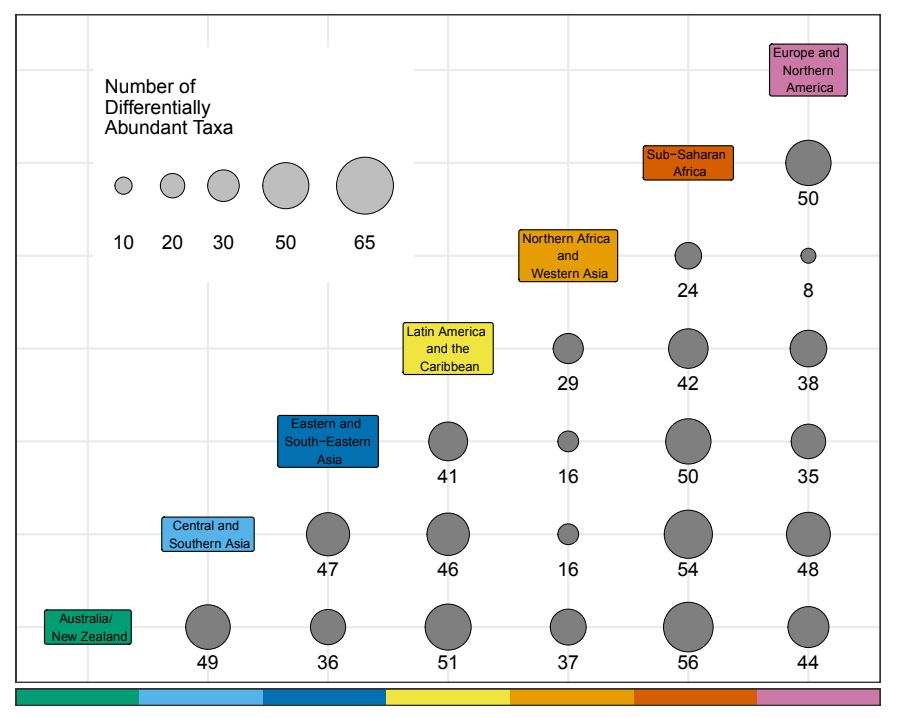
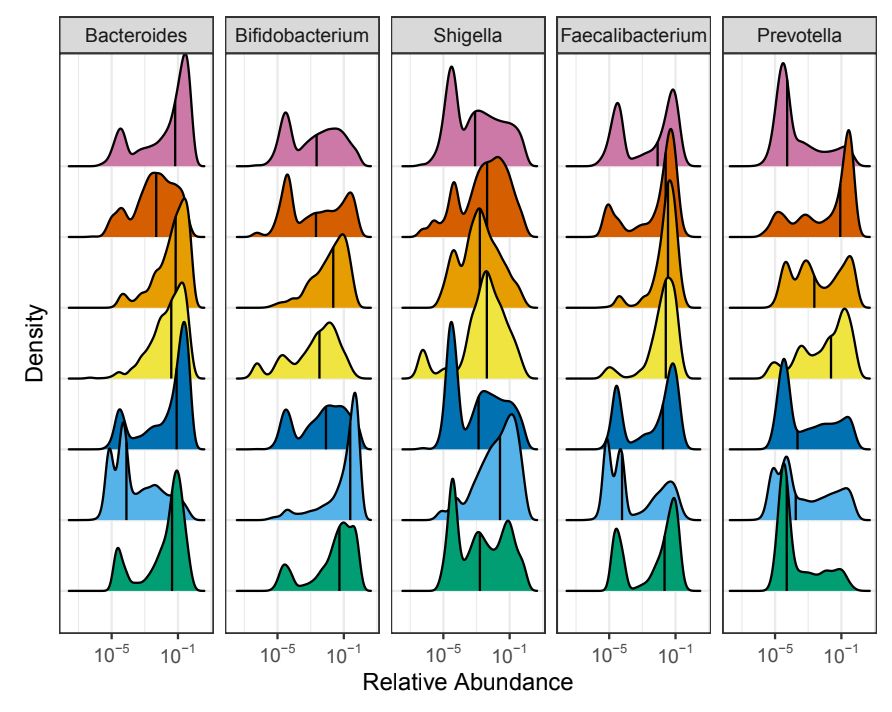
And specifically, we wanted to know what microbes were driving these large differences between world regions. We found that dozens of genera differ in abundance! Taxa like Bacteroides and Prevotella differ the most, but we found that every genus tested differed between at least two regions.
January 22, 2025 at 5:31 PM
And specifically, we wanted to know what microbes were driving these large differences between world regions. We found that dozens of genera differ in abundance! Taxa like Bacteroides and Prevotella differ the most, but we found that every genus tested differed between at least two regions.
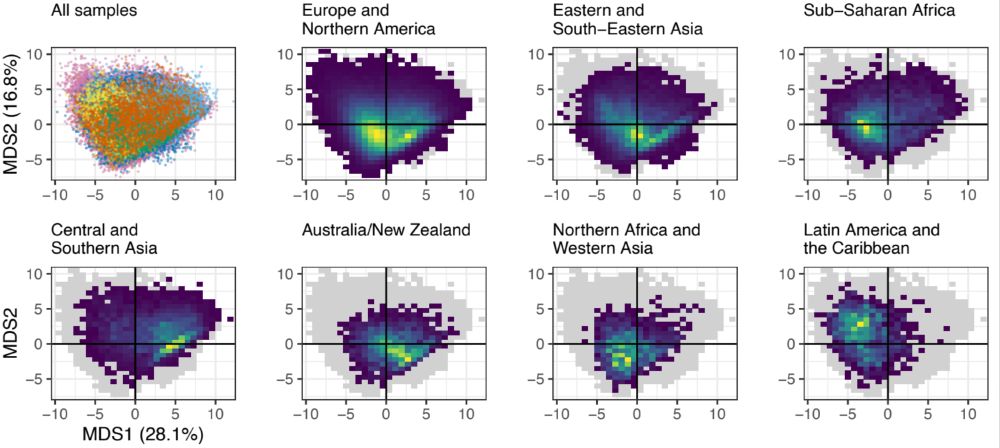
With sampling so unbalanced, are we missing a lot of information? To answer this, we wanted to see how much microbiomes differ between world regions. This big PCoA plot showed us that microbiomes from around the world have pretty drastic differences in composition.
January 22, 2025 at 5:31 PM
With sampling so unbalanced, are we missing a lot of information? To answer this, we wanted to see how much microbiomes differ between world regions. This big PCoA plot showed us that microbiomes from around the world have pretty drastic differences in composition.
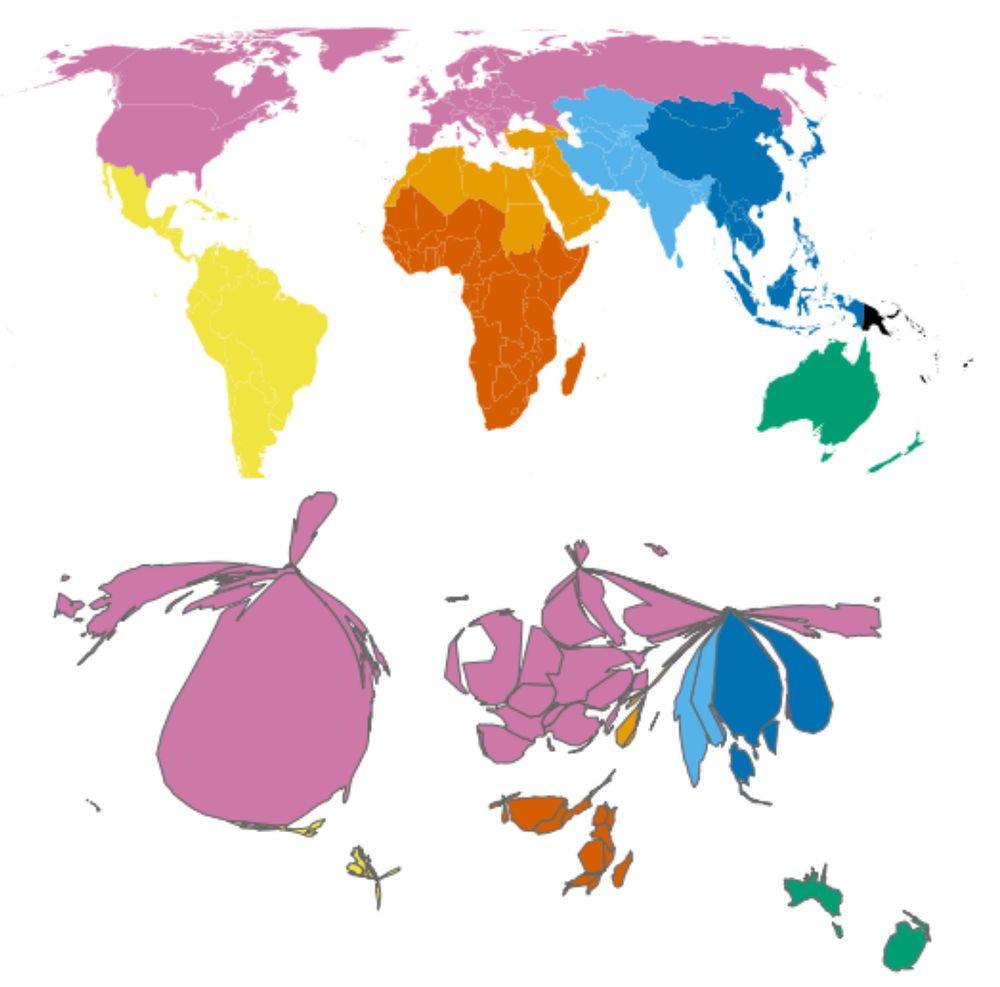
We previously found that microbiome sampling is unevenly distributed throughout the world (journals.plos.org/plosbiology/...), and we wanted to quantify just how uneven it is. Over half of our samples come from Europe/N. America, seen in this map that’s distorted to show sampling imbalance.
January 22, 2025 at 5:31 PM
We previously found that microbiome sampling is unevenly distributed throughout the world (journals.plos.org/plosbiology/...), and we wanted to quantify just how uneven it is. Over half of our samples come from Europe/N. America, seen in this map that’s distorted to show sampling imbalance.
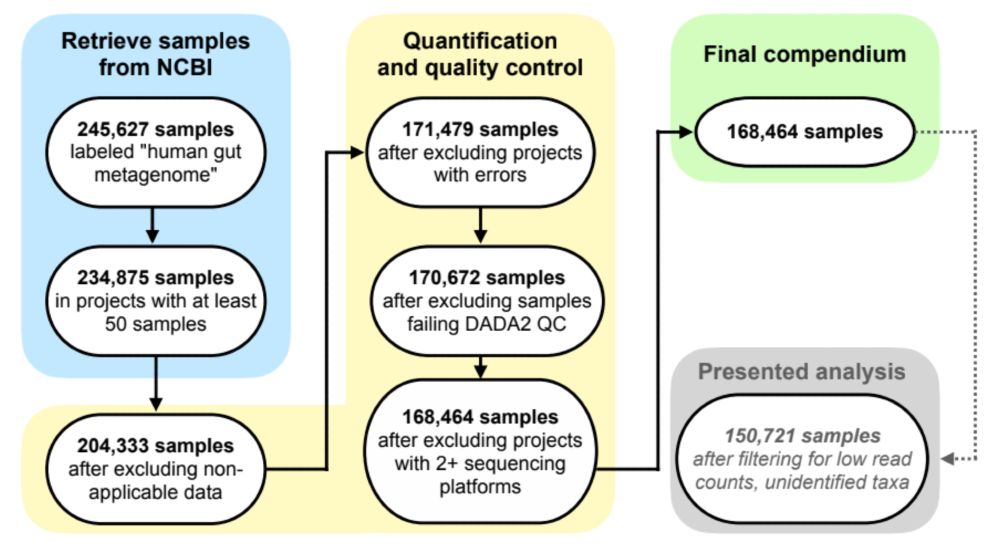
There’s tons of publicly available microbiome data, but this comes from 100s of studies that were processed separately, so we can’t directly compare results. We wanted to leverage this wealth of data, so we reprocessed raw data from 482 studies–that’s 5.7 terabases of data from all over the world!
January 22, 2025 at 5:31 PM
There’s tons of publicly available microbiome data, but this comes from 100s of studies that were processed separately, so we can’t directly compare results. We wanted to leverage this wealth of data, so we reprocessed raw data from 482 studies–that’s 5.7 terabases of data from all over the world!

