
I owe a massive debt of gratitude to the remarkable colleagues and mentors who have shaped my path, especially @ceesdekker.bsky.social !!

I owe a massive debt of gratitude to the remarkable colleagues and mentors who have shaped my path, especially @ceesdekker.bsky.social !!

Together, the two motifs account pretty much for the impact of the full NTR!! We hypothesize that connecting the two on one protein chain and some adjacent sequences modulate further.

Together, the two motifs account pretty much for the impact of the full NTR!! We hypothesize that connecting the two on one protein chain and some adjacent sequences modulate further.


- reduces cohesin's ATPase rate
- reduces LE initiation
- reduces the fraction of complete LE steps (in Magnetic Tweezers)
So these few amino acids alone already tinker quite a lot with cohesin!

- reduces cohesin's ATPase rate
- reduces LE initiation
- reduces the fraction of complete LE steps (in Magnetic Tweezers)
So these few amino acids alone already tinker quite a lot with cohesin!


but exactly how it pulls this off is a 'mechanistic mystery' (to cite @andersshansen.bsky.social, Nucleus, 2020).
We just preprinted 📜 a new study @biorxivpreprint.bsky.social that provides some answers to this enigma: www.biorxiv.org/content/10.1...
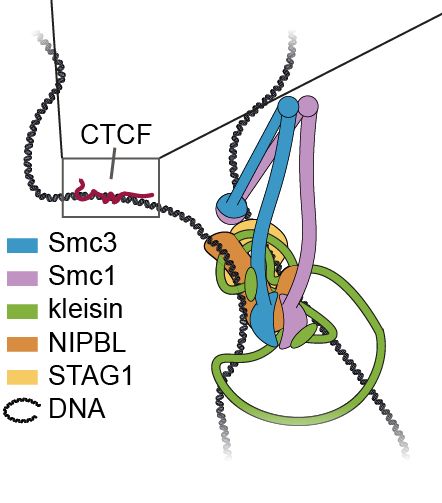
but exactly how it pulls this off is a 'mechanistic mystery' (to cite @andersshansen.bsky.social, Nucleus, 2020).
We just preprinted 📜 a new study @biorxivpreprint.bsky.social that provides some answers to this enigma: www.biorxiv.org/content/10.1...





Today we report on the dynamics of a fascinating class of molecular motors, so-called SMC proteins in @cp-cell.bsky.social
doi.org/10.1016/j.ce...

Today we report on the dynamics of a fascinating class of molecular motors, so-called SMC proteins in @cp-cell.bsky.social
doi.org/10.1016/j.ce...

Excitingly, this quantity appears to be conserved among cohesin, condensin, and SMC5/6, the 3 major SMCs in Eukaryotes

Excitingly, this quantity appears to be conserved among cohesin, condensin, and SMC5/6, the 3 major SMCs in Eukaryotes

We have been trying to figure out the mechanistic mysteries about SMC motor proteins. They extrude our genome constantly into loops. We know that that's important. But how it works, is a matter of intense debate! Today, we report a piece of the puzzle:

We have been trying to figure out the mechanistic mysteries about SMC motor proteins. They extrude our genome constantly into loops. We know that that's important. But how it works, is a matter of intense debate! Today, we report a piece of the puzzle:


