
Weekly symptoms of infection doesn't sound particularly appealing to me.
www.newscientist.com/article/2492...

Weekly symptoms of infection doesn't sound particularly appealing to me.
www.newscientist.com/article/2492...

Did Cold War virologists accidentally alter the evolution of a ubiquitous virus?
shorturl.at/HSke3

Did Cold War virologists accidentally alter the evolution of a ubiquitous virus?
shorturl.at/HSke3
Explore adaptable tools, resources, and projects developed in our lab using GLUE:
github.com/giffordlabcv...
#Genomics #Bioinformatics #OpenScience

Explore adaptable tools, resources, and projects developed in our lab using GLUE:
github.com/giffordlabcv...
#Genomics #Bioinformatics #OpenScience
On this evidence, the prospects for the promised HIV-1 vaccine and/or cure don't look particularly great.

On this evidence, the prospects for the promised HIV-1 vaccine and/or cure don't look particularly great.
The vast difference in the number of publications for SARS-CoV-2 compared to other viruses like Ebola, Dengue, and Influenza A is striking.
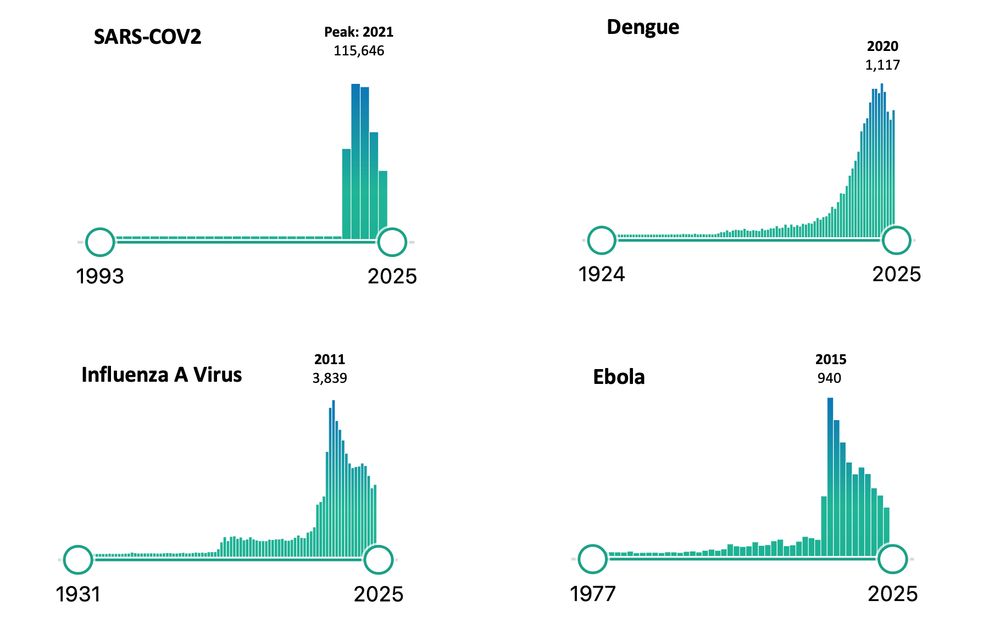
The vast difference in the number of publications for SARS-CoV-2 compared to other viruses like Ebola, Dengue, and Influenza A is striking.
GLUE's module system allows export of alignment partitions, based on reference sequence coordinates.
For example here we export a phosphoprotein alignment for HMPV (lineage B2).

GLUE's module system allows export of alignment partitions, based on reference sequence coordinates.
For example here we export a phosphoprotein alignment for HMPV (lineage B2).
Additionally, all sequences contained in HMPV-GLUE have been typed using MLCA.

Additionally, all sequences contained in HMPV-GLUE have been typed using MLCA.
With the recent rise in HMPV cases in China, this tool offers timely support for the scientific community.
github.com/giffordlabcv...

With the recent rise in HMPV cases in China, this tool offers timely support for the scientific community.
github.com/giffordlabcv...

