
Postdoc at the Theoretical Chemistry Group
Aix-Marseille Université
Molecular Dynamics | QM/MM | Quantum Mechanics | Biomolecules | Photophysics | Machine Learning
#compchem #stem #researcher #qmmm

github.com/Lemkul-Lab/g...

github.com/Lemkul-Lab/g...
We report the parametrization of the MST model for the prediction of hydration free energies based on ddCOSMO strategy. We also introduce several novelties in MST for both ddCOSMO and IEFPCM.
onlinelibrary.wiley.com/doi/10.1002/...
#compchem
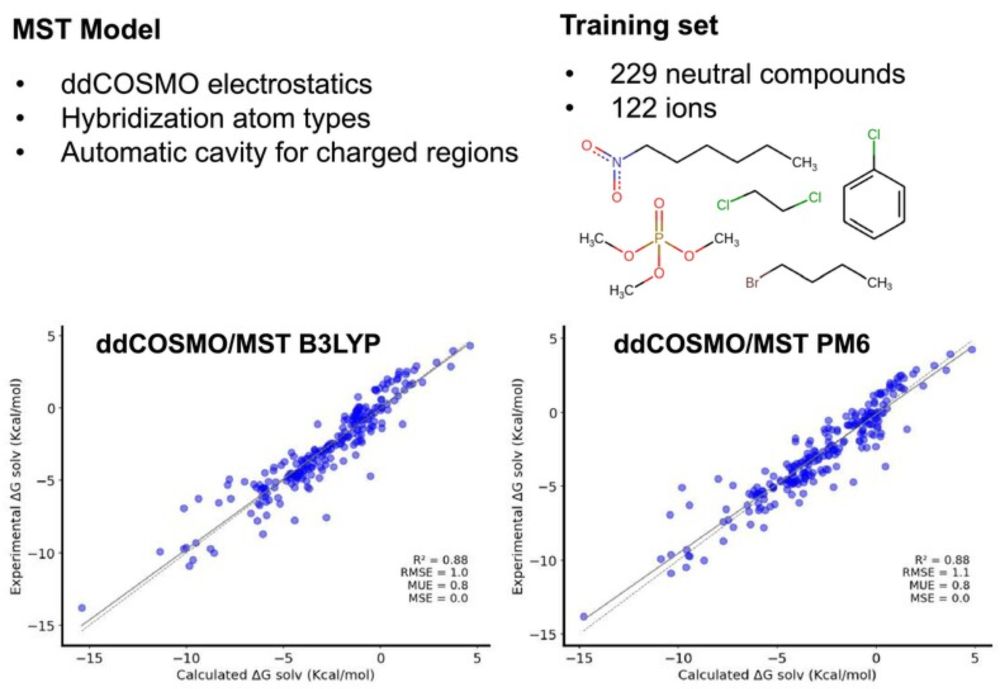
We report the parametrization of the MST model for the prediction of hydration free energies based on ddCOSMO strategy. We also introduce several novelties in MST for both ddCOSMO and IEFPCM.
onlinelibrary.wiley.com/doi/10.1002/...
#compchem

