
t.ly/OA9fS
Thank you for your interest in our work! (8/n)
t.ly/OA9fS
Thank you for your interest in our work! (8/n)
t.ly/S3JUv

t.ly/S3JUv


Specificity of 96.90%, discovering as well new alterations beyond the original scope of analysis within #TCGA. (5/n)

Specificity of 96.90%, discovering as well new alterations beyond the original scope of analysis within #TCGA. (5/n)
This iterative approach ensured diverse mutation representation, improving model robustness. (4/n)

This iterative approach ensured diverse mutation representation, improving model robustness. (4/n)
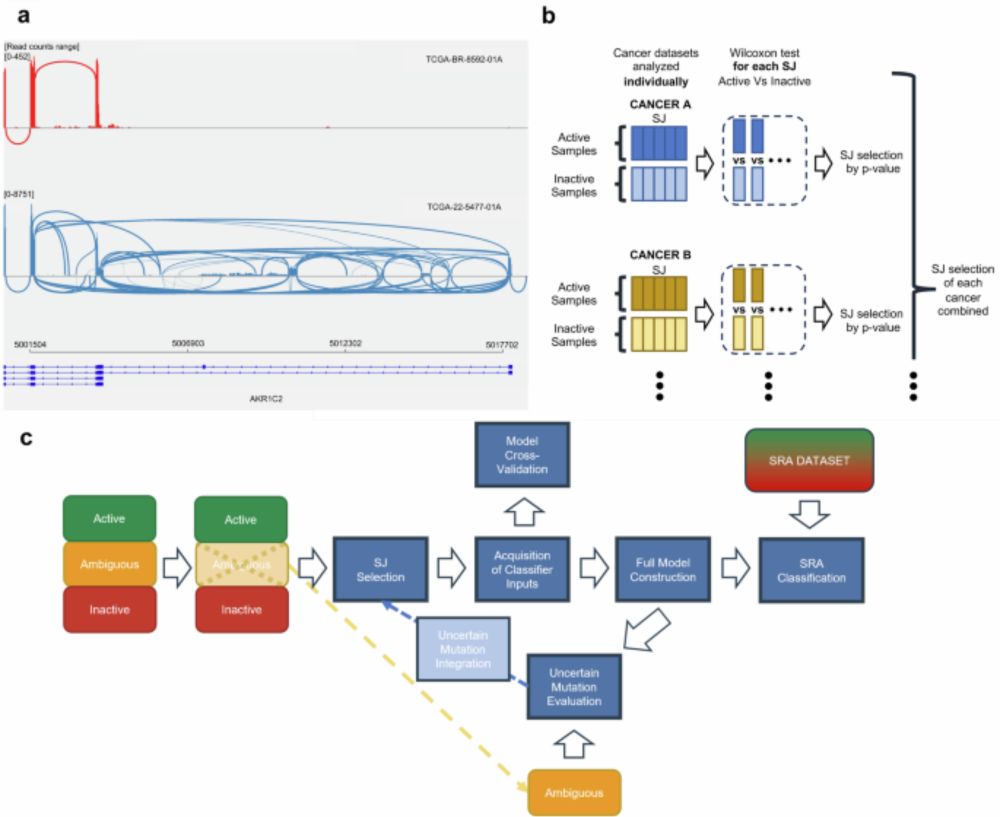
It focuses on mutations impacting pathway activity and leverages splicing junction signatures as biomarkers. (3/n)


It focuses on mutations impacting pathway activity and leverages splicing junction signatures as biomarkers. (3/n)
We explored a novel way to detect these activations using aberrant splicing junctions. (2/n)

We explored a novel way to detect these activations using aberrant splicing junctions. (2/n)

