Postdoctoral researcher | ImVA-HB / IDMIT @cea.fr | @yersiniaunit.bsky.social @pasteur.fr.
@csavin.bsky.social @julmini.bsky.social
@yersiniaunit.bsky.social @pasteur.fr @chrudetours.bsky.social @univparissaclay.bsky.social @cea.fr @ap-hp.bsky.social @natureportfolio.nature.com @natcomms.nature.com
@csavin.bsky.social @julmini.bsky.social
@yersiniaunit.bsky.social @pasteur.fr @chrudetours.bsky.social @univparissaclay.bsky.social @cea.fr @ap-hp.bsky.social @natureportfolio.nature.com @natcomms.nature.com
💊 A fascinating case of in-host bacterial evolution — and a warning about long-term antibiotic exposure.
💊 A fascinating case of in-host bacterial evolution — and a warning about long-term antibiotic exposure.
🚫 We also identified positive selection signals:
- a truncation in the phosphate transporter PitA which probably impairs its activity
- an amino acid substitution interfering with the active site and diminishing the catalytic activity of the acyl-CoA thioesterase II TesB

🚫 We also identified positive selection signals:
- a truncation in the phosphate transporter PitA which probably impairs its activity
- an amino acid substitution interfering with the active site and diminishing the catalytic activity of the acyl-CoA thioesterase II TesB
🍔 Omics analysis also unraveled:
- a progressive genome reduction
- metabolic changes in amino acid and carbon utilization
- a disturbed stringent response
which could contribute to the observed slower growth and antibiotic tolerance preceding resistance.

🍔 Omics analysis also unraveled:
- a progressive genome reduction
- metabolic changes in amino acid and carbon utilization
- a disturbed stringent response
which could contribute to the observed slower growth and antibiotic tolerance preceding resistance.
🧪 By analysing Ye.1 to Ye.4 genomes and proteomes, we could identify:
- a novel gyrA mutation leading to quinolone resistance
- ceftriaxone resistance via OmpF loss and β-lactamase activity
🧪 By analysing Ye.1 to Ye.4 genomes and proteomes, we could identify:
- a novel gyrA mutation leading to quinolone resistance
- ceftriaxone resistance via OmpF loss and β-lactamase activity
🧫 Ye.1 to Ye.4 belong to the Ye genotype 4 (bioserotype 4/O:3) and a form a unique clade separated from other Ye strains, strongly suggesting that the patient suffered from a chronic infection due to a unique evolving Ye 4/O:3 strain.

🧫 Ye.1 to Ye.4 belong to the Ye genotype 4 (bioserotype 4/O:3) and a form a unique clade separated from other Ye strains, strongly suggesting that the patient suffered from a chronic infection due to a unique evolving Ye 4/O:3 strain.
🏥 Thanks to the National Reference Center for Plague and other Yersiniosis in @pasteur.fr, we collected 4 clinical isolates — Ye.1 to Ye.4, which evolved antibiotics tolerance and resistance — from a rare chronic infection in an elderly patient from @chrudetours.bsky.social.
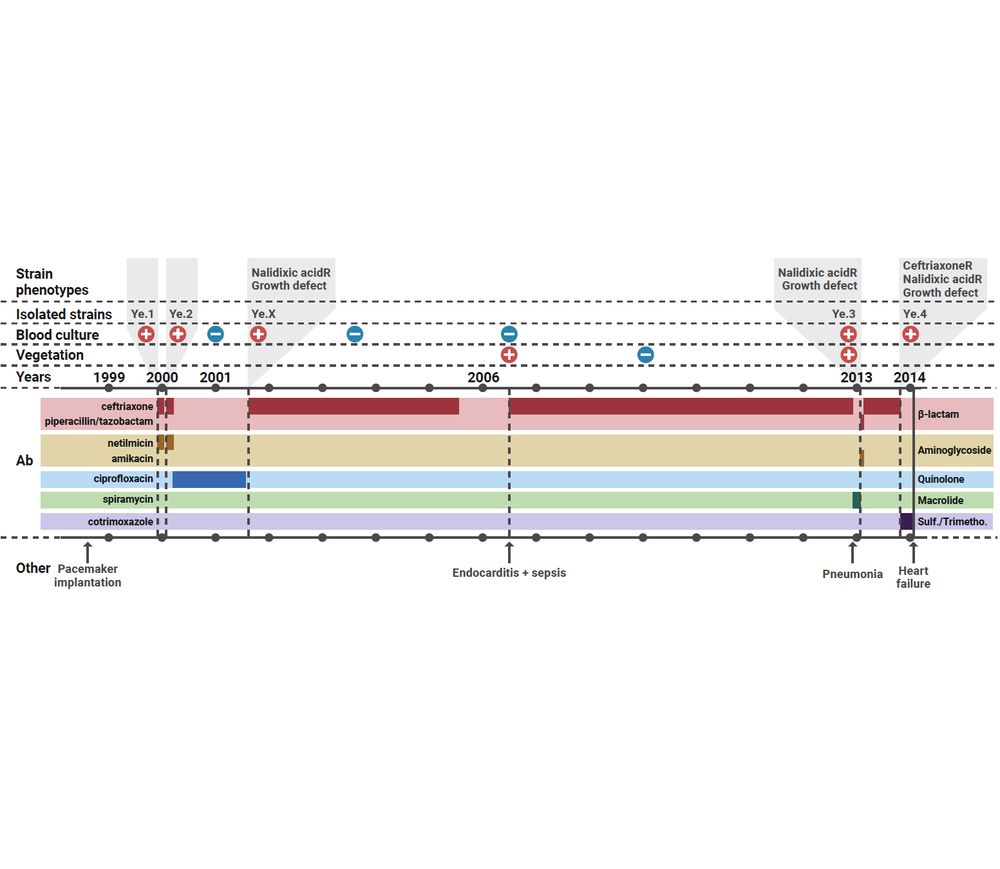
🏥 Thanks to the National Reference Center for Plague and other Yersiniosis in @pasteur.fr, we collected 4 clinical isolates — Ye.1 to Ye.4, which evolved antibiotics tolerance and resistance — from a rare chronic infection in an elderly patient from @chrudetours.bsky.social.
www.youtube.com/watch?v=o8nf...

www.youtube.com/watch?v=o8nf...
☠️ Curious of what happened to Yersinia pestis, the bacterium responsible for the Black Death, throughout plague pandemics?
Check out our new study 👇
www.science.org/doi/10.1126/...

@ravneetksidhu.bsky.social @gmasfiol.bsky.social @bbolker.bsky.social @yersiniaunit.bsky.social
#ScienceResearch
@ravneetksidhu.bsky.social @gmasfiol.bsky.social @bbolker.bsky.social @yersiniaunit.bsky.social
#ScienceResearch
Our study highlights the importance of reservoir dynamics and structures to better understand virulence evolution of a zoonotic pathogen.
Our study highlights the importance of reservoir dynamics and structures to better understand virulence evolution of a zoonotic pathogen.

