
Paula Ramiro-Martínez
@paularamiro.bsky.social
Predoctoral researcher at evodynamics lab in Madrid.
Bacterial and plasmid evolution 🫧🧬💻
Bacterial and plasmid evolution 🫧🧬💻
Thirty days of evolution… totally worth it! 🥹

July 22, 2025 at 10:20 AM
Thirty days of evolution… totally worth it! 🥹
Do natural plasmids show the same results?
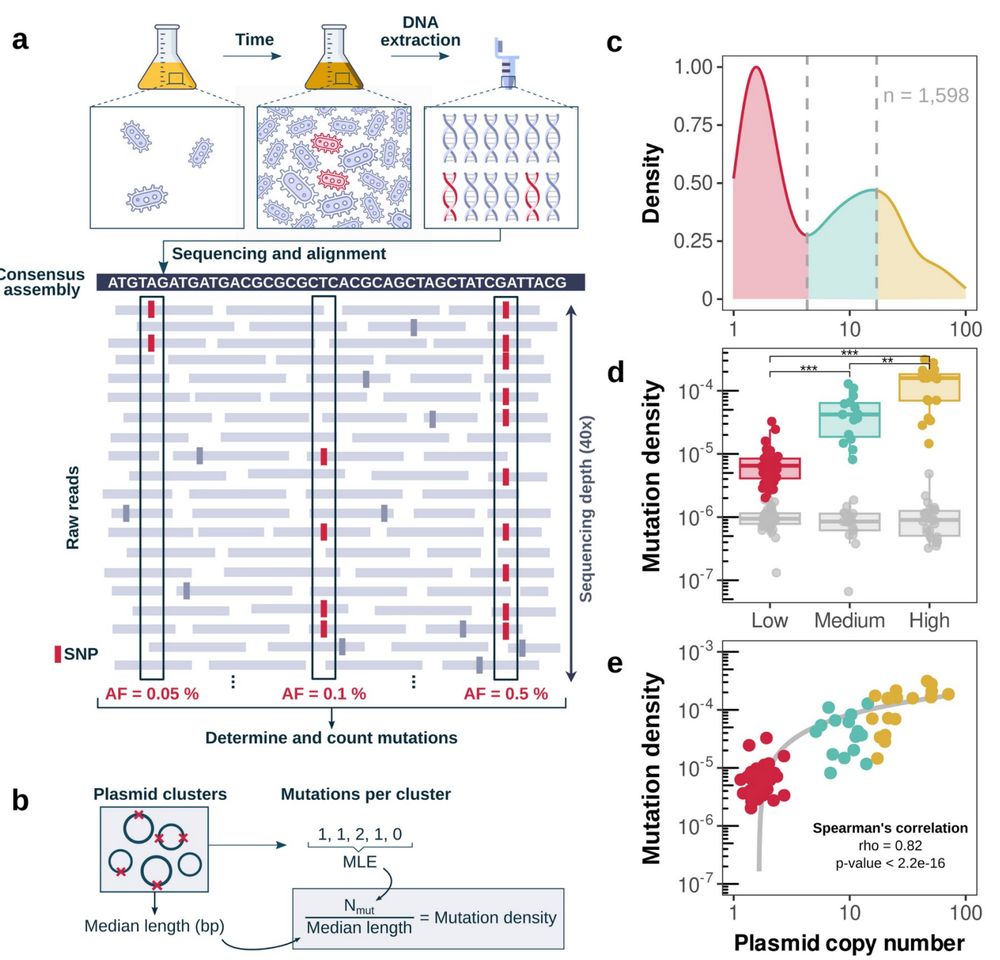
We analyzed 1,598 plasmids (NCBI) 💻 to detect mutations arising during culture growth by comparing sequencing reads to consensus assemblies.
High-copy plasmids exhibited significantly higher mutation rates, just like theory (and experiments!) predicted!
We analyzed 1,598 plasmids (NCBI) 💻 to detect mutations arising during culture growth by comparing sequencing reads to consensus assemblies.
High-copy plasmids exhibited significantly higher mutation rates, just like theory (and experiments!) predicted!
July 22, 2025 at 9:59 AM
Do natural plasmids show the same results?
We analyzed 1,598 plasmids (NCBI) 💻 to detect mutations arising during culture growth by comparing sequencing reads to consensus assemblies.
High-copy plasmids exhibited significantly higher mutation rates, just like theory (and experiments!) predicted!
We analyzed 1,598 plasmids (NCBI) 💻 to detect mutations arising during culture growth by comparing sequencing reads to consensus assemblies.
High-copy plasmids exhibited significantly higher mutation rates, just like theory (and experiments!) predicted!
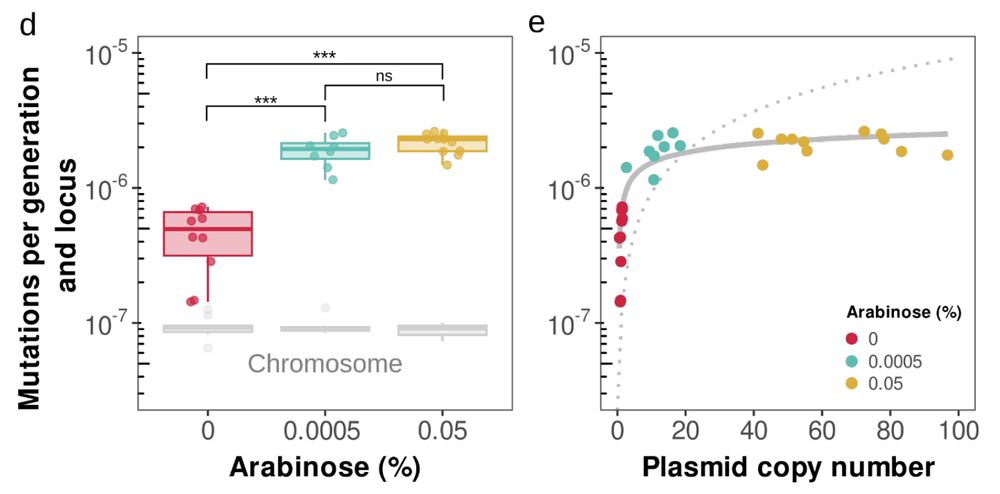
Experiments confirm it: plasmid mutation rate rises logarithmically with copy number 🚀
At more copies, mutation supply dominates drift! 💥
At more copies, mutation supply dominates drift! 💥
July 22, 2025 at 9:55 AM
Experiments confirm it: plasmid mutation rate rises logarithmically with copy number 🚀
At more copies, mutation supply dominates drift! 💥
At more copies, mutation supply dominates drift! 💥
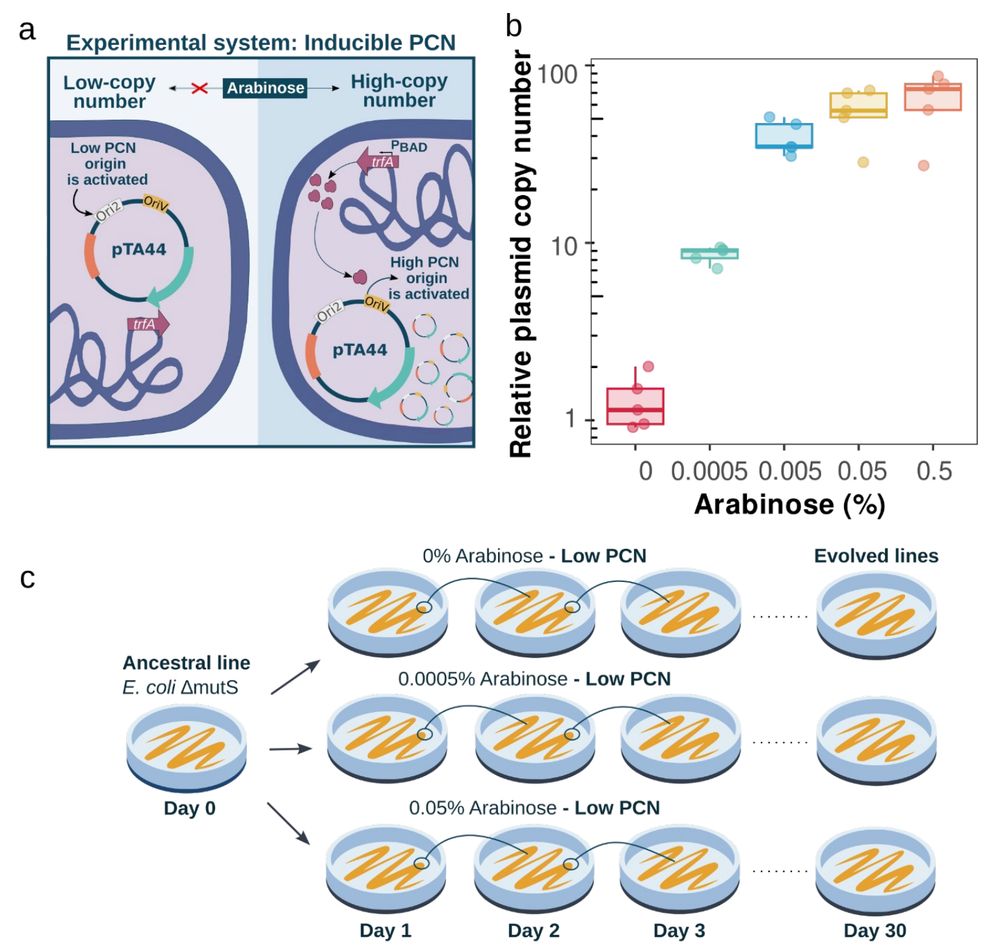
We conducted a mutation accumulation experiment to test our predictions using a plasmid with a tunable copy number 🫧
We established three PCN conditions: low (~1), medium (~10), and high (~60 copies). Lines were evolved for 30 days under severe bottlenecks.
We established three PCN conditions: low (~1), medium (~10), and high (~60 copies). Lines were evolved for 30 days under severe bottlenecks.
July 22, 2025 at 9:51 AM
We conducted a mutation accumulation experiment to test our predictions using a plasmid with a tunable copy number 🫧
We established three PCN conditions: low (~1), medium (~10), and high (~60 copies). Lines were evolved for 30 days under severe bottlenecks.
We established three PCN conditions: low (~1), medium (~10), and high (~60 copies). Lines were evolved for 30 days under severe bottlenecks.
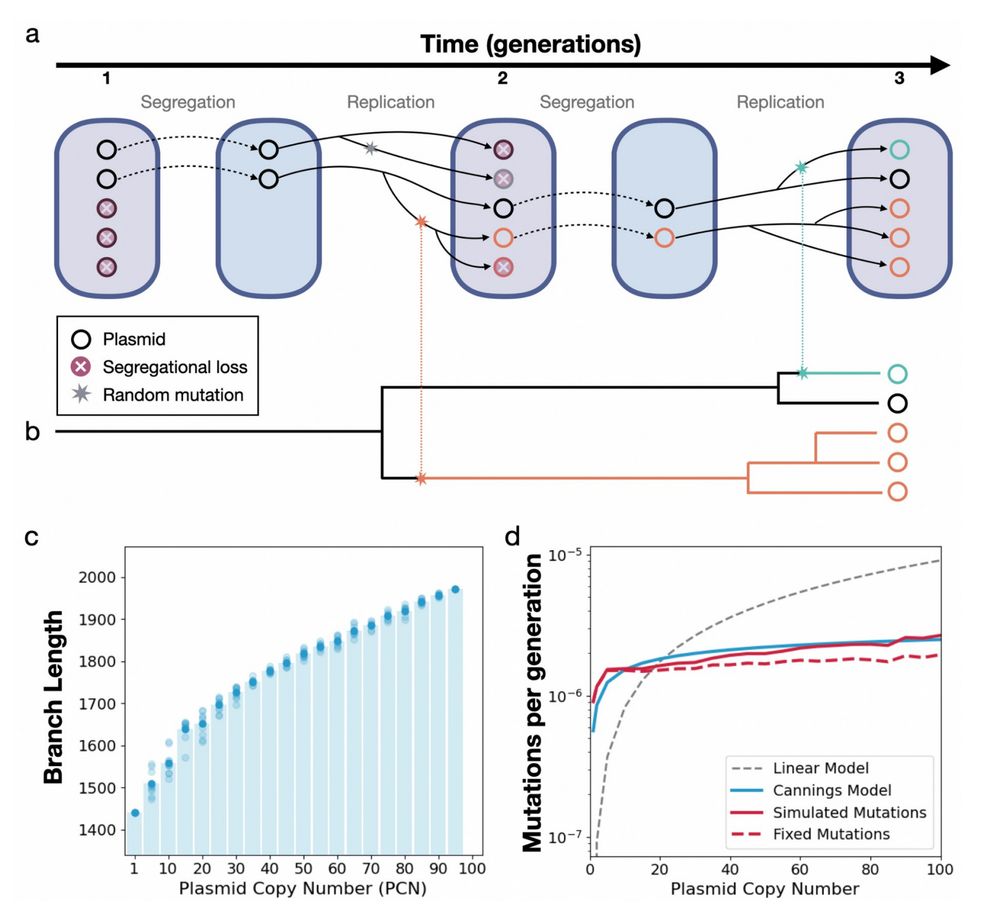
We modelled plasmid inheritance using a Cannings model and validated it with computational simulations 🧫 Each plasmid copy is like an evolutionary unit, randomly split at division, then copied.
Prediction: the mutation rate rises with PCN, but with diminishing returns, increasing logarithmically.
Prediction: the mutation rate rises with PCN, but with diminishing returns, increasing logarithmically.
July 22, 2025 at 9:47 AM
We modelled plasmid inheritance using a Cannings model and validated it with computational simulations 🧫 Each plasmid copy is like an evolutionary unit, randomly split at division, then copied.
Prediction: the mutation rate rises with PCN, but with diminishing returns, increasing logarithmically.
Prediction: the mutation rate rises with PCN, but with diminishing returns, increasing logarithmically.
Plasmids face opposing evolutionary forces:
🔹 More copies = more chances to mutate 📈
🔹 But random segregation = drift, purging mutations 📉
So mutation supply ↑ with PCN, but the probability of fixation ↓
Which force dominates: the boost in mutation rate or the loss from segregational drift?
🔹 More copies = more chances to mutate 📈
🔹 But random segregation = drift, purging mutations 📉
So mutation supply ↑ with PCN, but the probability of fixation ↓
Which force dominates: the boost in mutation rate or the loss from segregational drift?
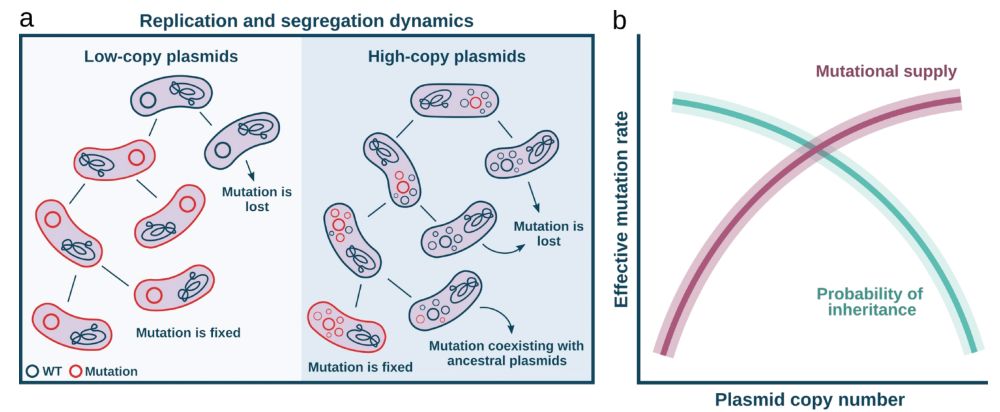
July 22, 2025 at 9:42 AM
Plasmids face opposing evolutionary forces:
🔹 More copies = more chances to mutate 📈
🔹 But random segregation = drift, purging mutations 📉
So mutation supply ↑ with PCN, but the probability of fixation ↓
Which force dominates: the boost in mutation rate or the loss from segregational drift?
🔹 More copies = more chances to mutate 📈
🔹 But random segregation = drift, purging mutations 📉
So mutation supply ↑ with PCN, but the probability of fixation ↓
Which force dominates: the boost in mutation rate or the loss from segregational drift?
We discovered a universal scaling law linking copy number and plasmid size, which can be used to predict the PCN of your favorite plasmid! 6/7
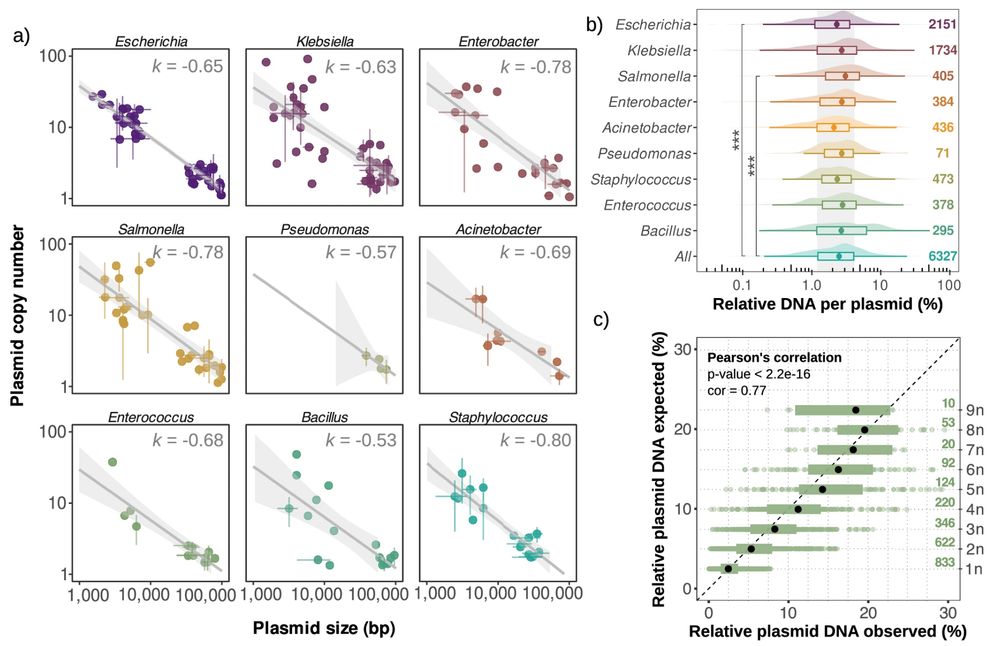
July 2, 2025 at 10:12 AM
We discovered a universal scaling law linking copy number and plasmid size, which can be used to predict the PCN of your favorite plasmid! 6/7
Furthermore, we investigate the interactions between different replicons in co-integrated plasmids and coin the concept of replicon dominance. 5/7
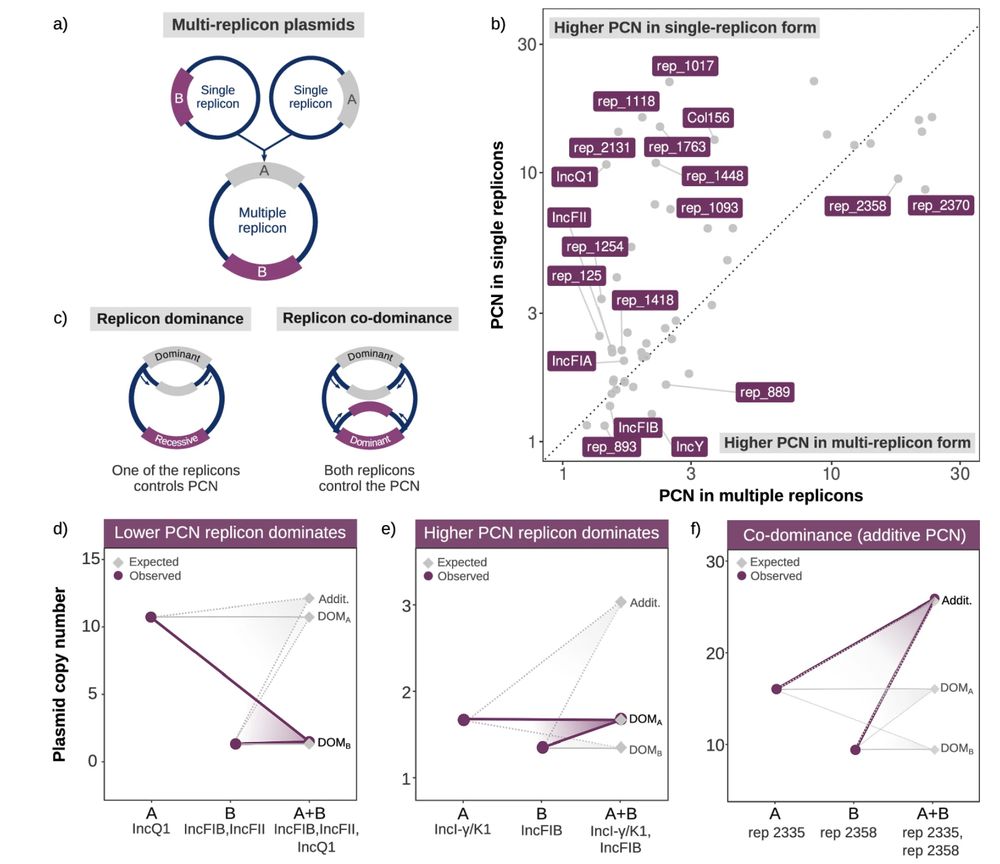
July 2, 2025 at 10:11 AM
Furthermore, we investigate the interactions between different replicons in co-integrated plasmids and coin the concept of replicon dominance. 5/7
We found that variability in copy number is closely associated with plasmid lifestyles, with small plasmids being more plastic in their copy number. 4/7
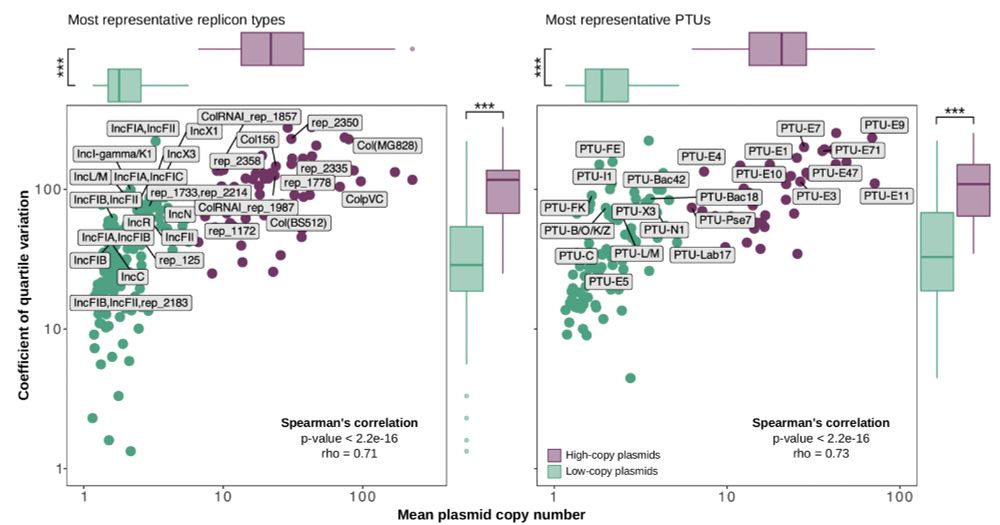
July 2, 2025 at 10:10 AM
We found that variability in copy number is closely associated with plasmid lifestyles, with small plasmids being more plastic in their copy number. 4/7
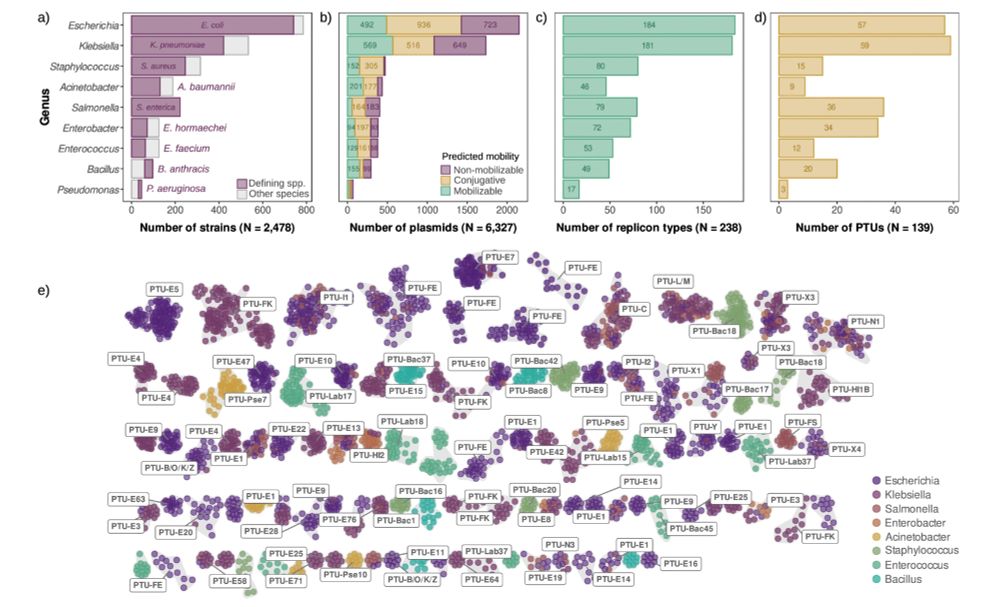
Our results revealed that although the copy number of natural plasmids is extremely diverse, it is robust against changes in genetic content and host 🦠. 3/7
July 2, 2025 at 10:09 AM
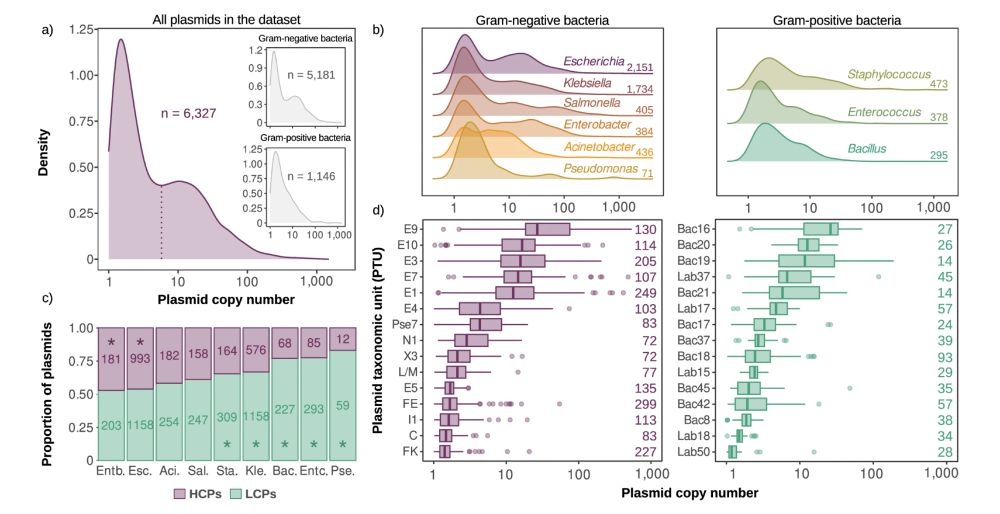
Our results revealed that although the copy number of natural plasmids is extremely diverse, it is robust against changes in genetic content and host 🦠. 3/7
First, we used DNA sequencing data to calculate and analyze the copy numbers of >6300 plasmids from different bacterial genera 💻. 2/7
July 2, 2025 at 10:08 AM
First, we used DNA sequencing data to calculate and analyze the copy numbers of >6300 plasmids from different bacterial genera 💻. 2/7

