Pam Engelberts
@pam-engelberts.bsky.social
Post doc @cmrqut.bsky.social with a love for imaging, baking, and nature
Part of BrisJAMS @brisjams.bsky.social and ISME ECSC @isme-microbes.bsky.social
Part of BrisJAMS @brisjams.bsky.social and ISME ECSC @isme-microbes.bsky.social
Thanks Steve! It was great working with you on the BrisJAMS committee 🙂
September 15, 2025 at 10:11 AM
Thanks Steve! It was great working with you on the BrisJAMS committee 🙂
Thank you Courtney! GenomeFISH should work on roots as well, as long as you can get the genomes of the microbes of interest for probe generation :)
July 17, 2025 at 11:10 PM
Thank you Courtney! GenomeFISH should work on roots as well, as long as you can get the genomes of the microbes of interest for probe generation :)
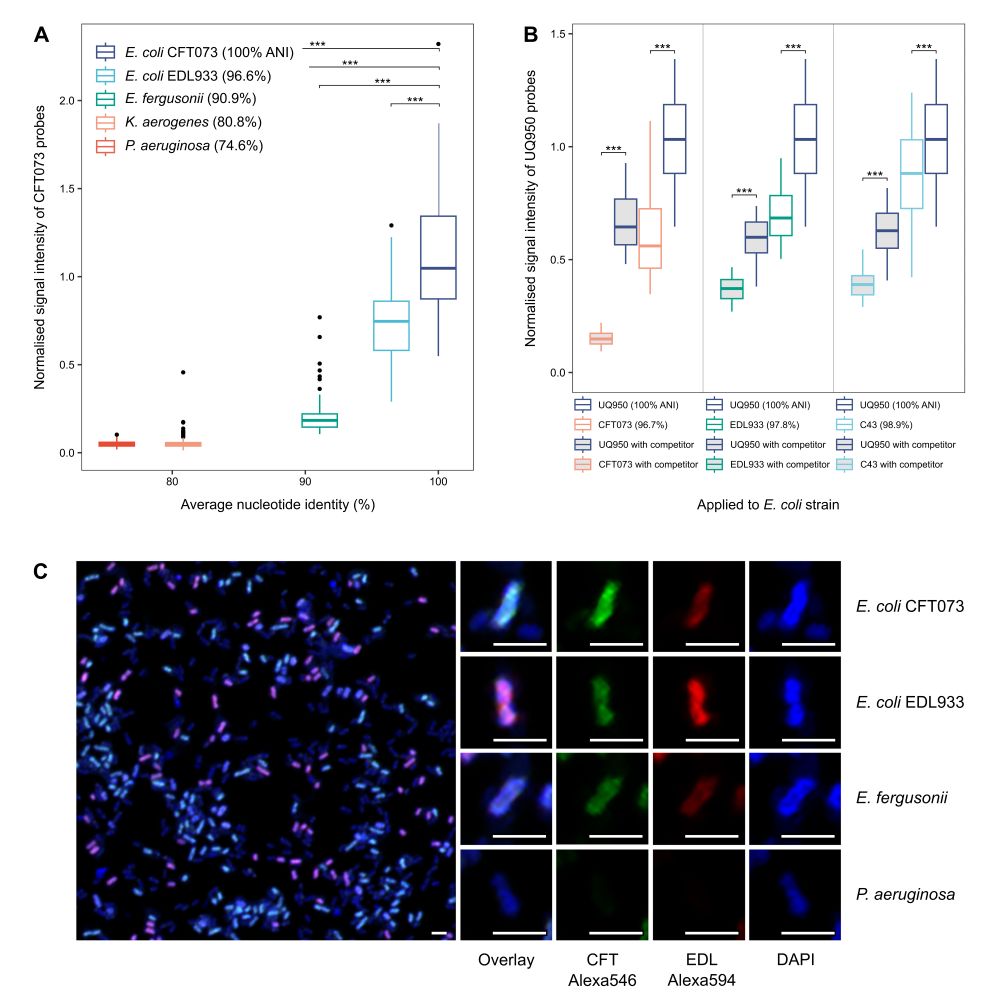
In the target population, the GenomeFISH and DAPI signal are positively correlated, resulting in a normalised signal intensity of ~1 when dividing the GenomeFISH signal by the DAPI signal. For closely related strains, this value drops below 1, as the GenomeFISH signal reduces in strength.
(2/2)
(2/2)
July 8, 2025 at 10:58 PM
In the target population, the GenomeFISH and DAPI signal are positively correlated, resulting in a normalised signal intensity of ~1 when dividing the GenomeFISH signal by the DAPI signal. For closely related strains, this value drops below 1, as the GenomeFISH signal reduces in strength.
(2/2)
(2/2)
Thanks Nicholas! We normalise the GenomeFISH signal to the DAPI signal. As genome copy number and image plane orientation can all influence the signal strength in the target population and probes will also bind to closely related strains, normalisation is key.
(1/2)
(1/2)
July 8, 2025 at 10:56 PM
Thanks Nicholas! We normalise the GenomeFISH signal to the DAPI signal. As genome copy number and image plane orientation can all influence the signal strength in the target population and probes will also bind to closely related strains, normalisation is key.
(1/2)
(1/2)
No need for design :) GenomeFISH probes are generated from single amplified genomes or isolate genomes by simply fragmenting the entire genome and fluorescently labelling the fragments.
July 8, 2025 at 6:48 AM
No need for design :) GenomeFISH probes are generated from single amplified genomes or isolate genomes by simply fragmenting the entire genome and fluorescently labelling the fragments.
Given the high-throughput nature of GenomeFISH, its superior sensitivity, and its phylogenetic resolution, we hope it will become widely used for the visualisation of complex microbial communities.
July 8, 2025 at 2:33 AM
Given the high-throughput nature of GenomeFISH, its superior sensitivity, and its phylogenetic resolution, we hope it will become widely used for the visualisation of complex microbial communities.
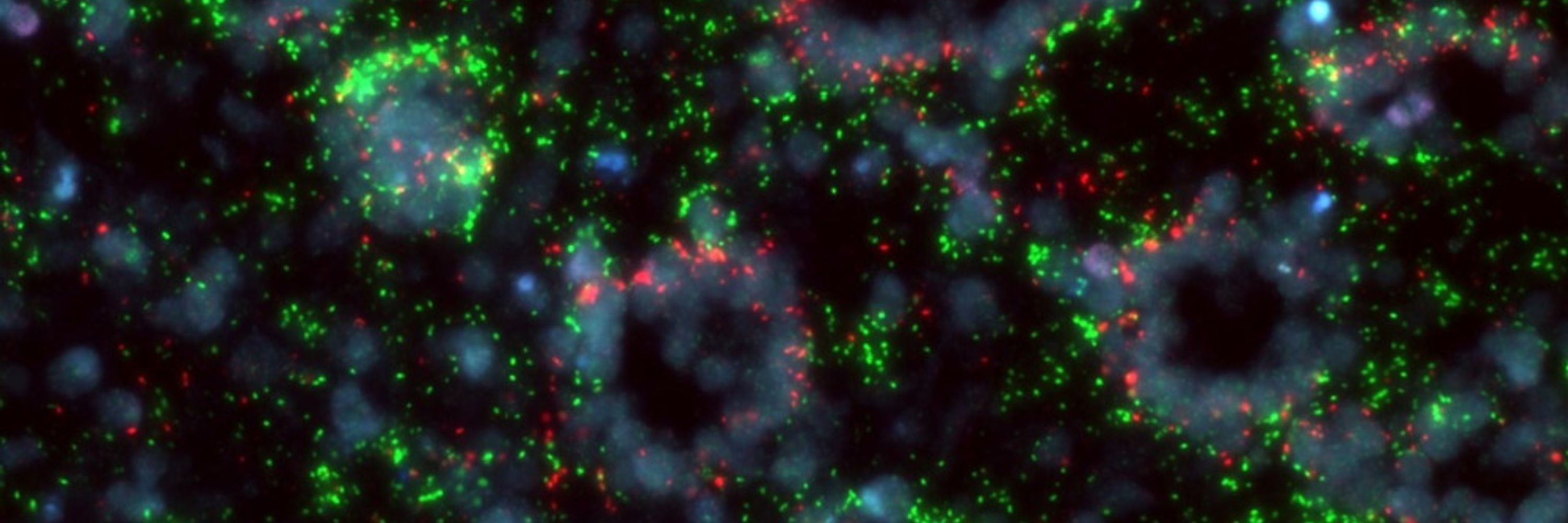
It also enables GenomeFISH to reach superior signal intensities compared to traditional FISH, allowing it to visualise microbial lineages that are challenging to visualise with traditional FISH, such as members of the Patescibacteria.
July 8, 2025 at 2:33 AM
It also enables GenomeFISH to reach superior signal intensities compared to traditional FISH, allowing it to visualise microbial lineages that are challenging to visualise with traditional FISH, such as members of the Patescibacteria.
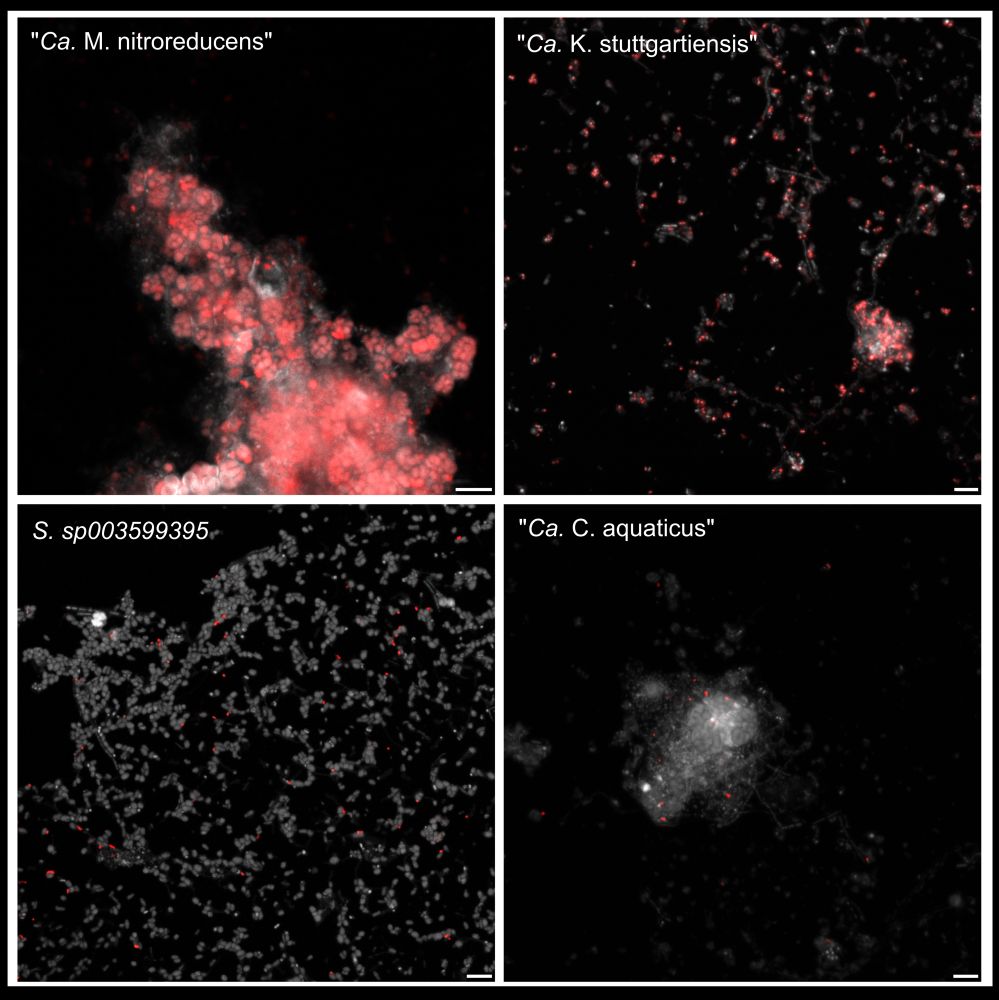
As a result, GenomeFISH can distinguish between strains with up to 99% ANI in mock communities and environmental samples.
July 8, 2025 at 2:32 AM
As a result, GenomeFISH can distinguish between strains with up to 99% ANI in mock communities and environmental samples.
By targeting the entire genome, GenomeFISH overcomes the inherent limitations of traditional FISH, circumventing the need for probe design and optimisation, while increasing sensitivity and specificity.
July 8, 2025 at 2:32 AM
By targeting the entire genome, GenomeFISH overcomes the inherent limitations of traditional FISH, circumventing the need for probe design and optimisation, while increasing sensitivity and specificity.
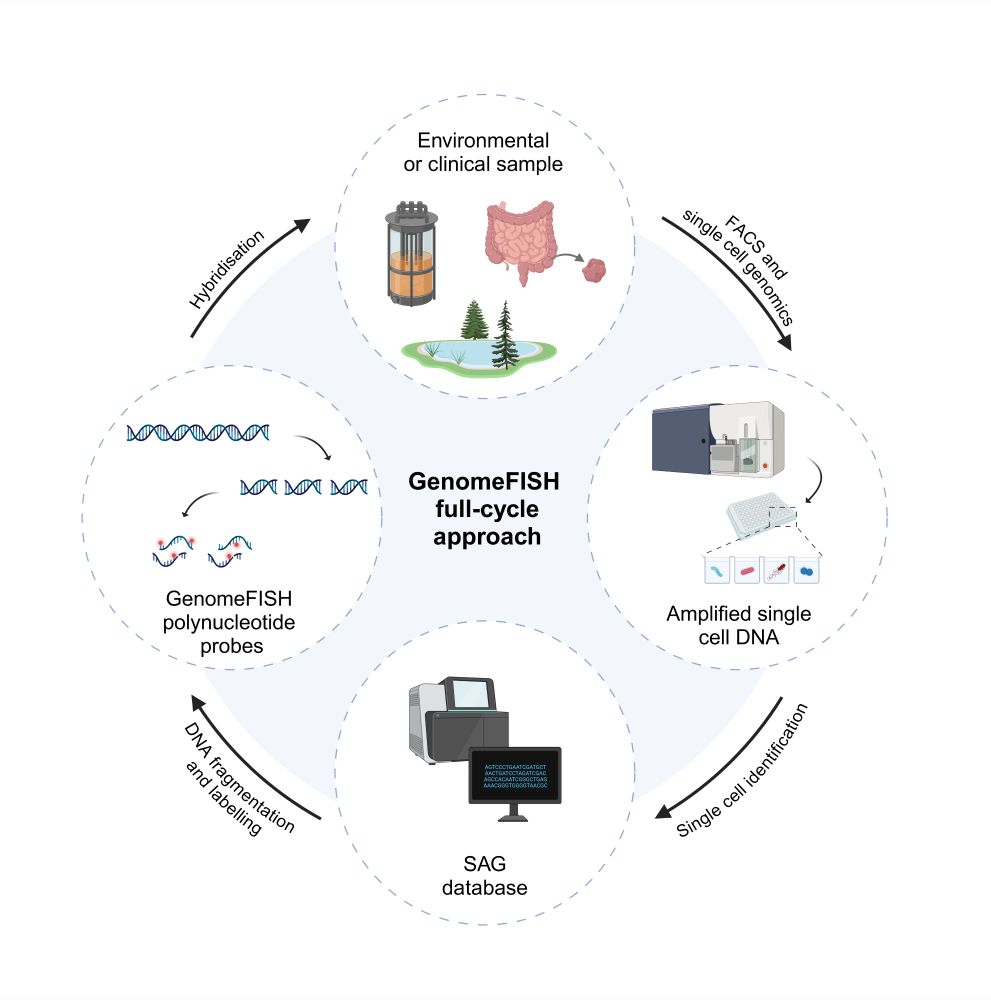
GenomeFISH probes can be rapidly generated from single cell genomes obtained from environmental and clinical samples, which are amplified, fragmented, fluorescently labelled, and hybridised to visualise the target microorganism in situ.
July 8, 2025 at 2:31 AM
GenomeFISH probes can be rapidly generated from single cell genomes obtained from environmental and clinical samples, which are amplified, fragmented, fluorescently labelled, and hybridised to visualise the target microorganism in situ.
GenomeFISH is an innovative, genome-based FISH approach that couples high-throughput single-cell genomics with whole-genome hybridisation to visualise microbial communities.
July 8, 2025 at 2:31 AM
GenomeFISH is an innovative, genome-based FISH approach that couples high-throughput single-cell genomics with whole-genome hybridisation to visualise microbial communities.

