





While oxbow makes legacy data more accessible, it is a good conduit to more general-purpose persistent storage.

While oxbow makes legacy data more accessible, it is a good conduit to more general-purpose persistent storage.
github.com/jmschrei/bpn...
bsky.app/profile/jmsc...
github.com/jmschrei/bpn...
bsky.app/profile/jmsc...
`pip install oxbow`
`pip install oxbow`
cfp.scipy.org/scipy2025/ta...
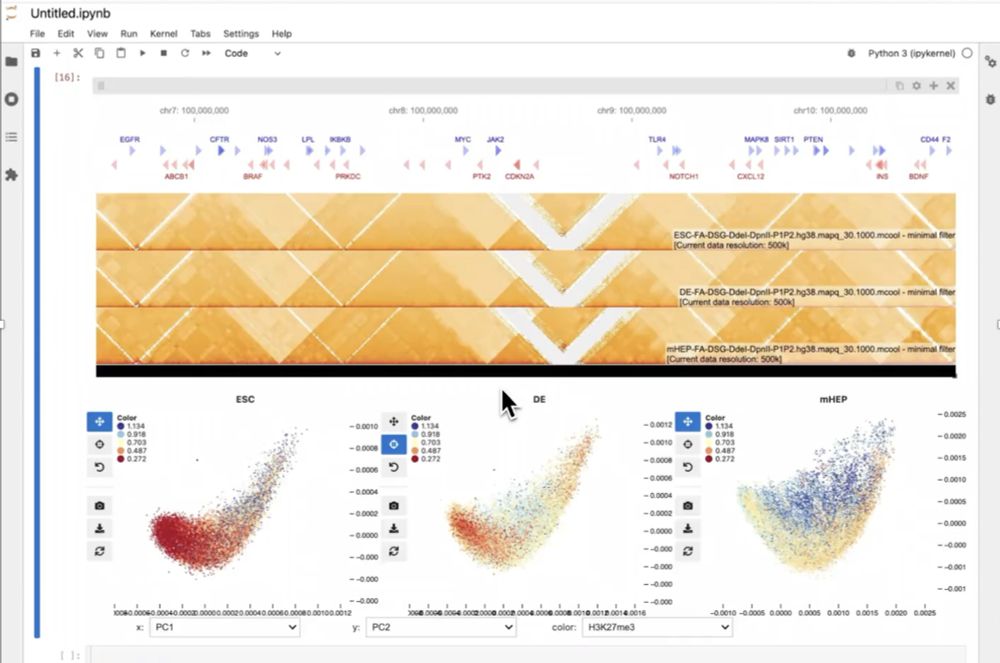
cfp.scipy.org/scipy2025/ta...
* Column projection and pushdown (parsing only the fields you need)
* Complex and nested field types (e.g. alignment tags, variant genotype call data, etc.)
* Genomic range-based queries via an index
* User-defined transports and file systems
* Column projection and pushdown (parsing only the fields you need)
* Complex and nested field types (e.g. alignment tags, variant genotype call data, etc.)
* Genomic range-based queries via an index
* User-defined transports and file systems
`pip install oxbow`
`pip install oxbow`
cfp.scipy.org/scipy2025/ta...

cfp.scipy.org/scipy2025/ta...
* Column projection and pushdown (parsing only the fields you need)
* Complex and nested field types (e.g. alignment tags, variant genotype call data, etc.)
* Genomic range-based queries via an index
* User-defined transports and file systems
* Column projection and pushdown (parsing only the fields you need)
* Complex and nested field types (e.g. alignment tags, variant genotype call data, etc.)
* Genomic range-based queries via an index
* User-defined transports and file systems

