
The FASTA format is now 40 years old (Pearson & Lipman) and any2fasta makes it easy for your scripts and pipelines that accept FASTA to also accept other formats, even if compressed! eg. .gbk.gz
#bioinformatiocs #microbiology #genomcs
github.com/tseemann/any...

The FASTA format is now 40 years old (Pearson & Lipman) and any2fasta makes it easy for your scripts and pipelines that accept FASTA to also accept other formats, even if compressed! eg. .gbk.gz
#bioinformatiocs #microbiology #genomcs
github.com/tseemann/any...
This is the last major release of Prokka. But don't be sad, because @oschwengers.bsky.social already has an excellent replacement called Bakta you can migrate to.
#bioinformatics #microbiology #genomics
github.com/tseemann/pro...
This is the last major release of Prokka. But don't be sad, because @oschwengers.bsky.social already has an excellent replacement called Bakta you can migrate to.
#bioinformatics #microbiology #genomics
github.com/tseemann/pro...
lh3.github.io/2025/09/11/a...

lh3.github.io/2025/09/11/a...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...

It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...

doi.org/10.1007/s111...

doi.org/10.1007/s111...
www.microbiologyresearch.org/content/jour...

www.microbiologyresearch.org/content/jour...
Excludons are pairs of overlapping genes that block each other’s expression (basically, reverse operons).
We built a tool to identify them in bacterial genomes using transcriptomic data, in an awesome collab led by Iñigo Lasa and Álvaro San Martín.
👇
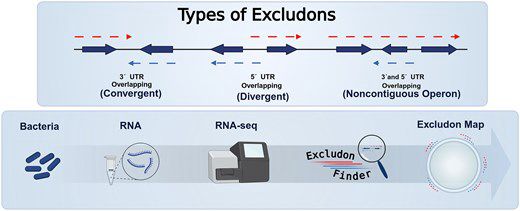
Excludons are pairs of overlapping genes that block each other’s expression (basically, reverse operons).
We built a tool to identify them in bacterial genomes using transcriptomic data, in an awesome collab led by Iñigo Lasa and Álvaro San Martín.
👇
Excludons are pairs of overlapping genes that block each other’s expression (basically, reverse operons).
We built a tool to identify them in bacterial genomes using transcriptomic data, in an awesome collab led by Iñigo Lasa and Álvaro San Martín.
👇

After spotting weird gene‐clustering and 'pangenome' quirks, I dug deep and found it’s not just one tool. It’s how we misuse and misassume how 'black-box' clustering and pangenome tools work.
#pangenomics #bioinformatics #geneclustering

After spotting weird gene‐clustering and 'pangenome' quirks, I dug deep and found it’s not just one tool. It’s how we misuse and misassume how 'black-box' clustering and pangenome tools work.
#pangenomics #bioinformatics #geneclustering
Actual title: Chromosomal capture of beneficial genes drives plasmids towards ecological redundancy.
Sensationalist title: Plasmids carry useless genes
academic.oup.com/ismej/articl...
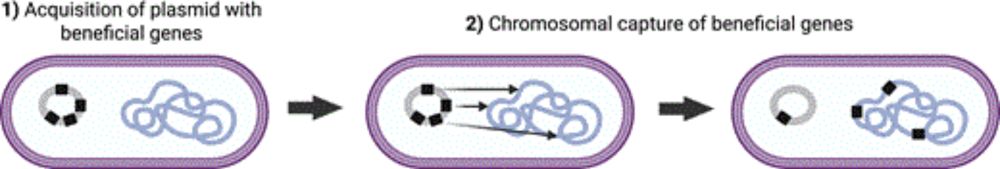
Actual title: Chromosomal capture of beneficial genes drives plasmids towards ecological redundancy.
Sensationalist title: Plasmids carry useless genes
academic.oup.com/ismej/articl...
(here the real paper: link.springer.com/article/10.1...)

(here the real paper: link.springer.com/article/10.1...)




