
Professor for Molecular Pathology in Neuro-Oncology, Neuropathologist at University Hospital Hamburg-Eppendorf
Research Interests: Proteomics, DNA-methylomics, Bioinformatics, Preclinical Models,
Digital Pathology
We present our new tool CCNV - now out in BMC Bioinformatics!
link.springer.com/article/10.1...

We present our new tool CCNV - now out in BMC Bioinformatics!
link.springer.com/article/10.1...
Meet BERT (our new algorithm Batch-Effect Reduction Trees) - details in #naturecommunications :
www.nature.com/articles/s41...
#transcriptomics #proteomics #metabolomics #dataanalysis #zmnh #ukeHamburg
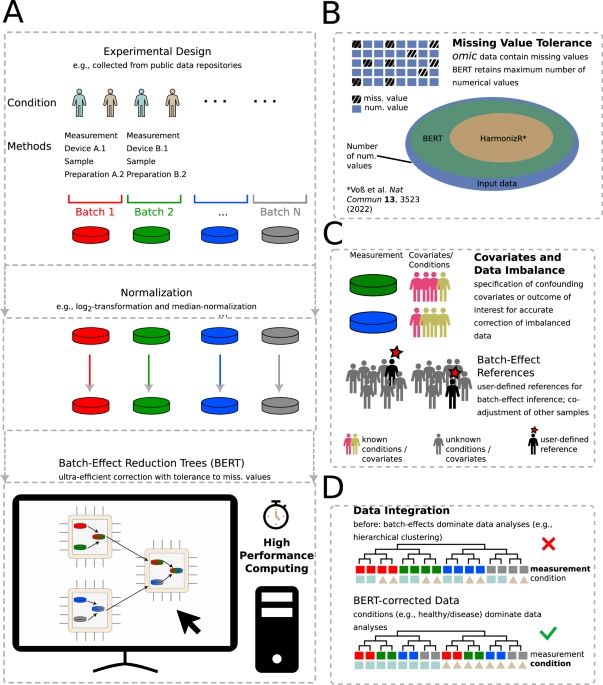
Meet BERT (our new algorithm Batch-Effect Reduction Trees) - details in #naturecommunications :
www.nature.com/articles/s41...
#transcriptomics #proteomics #metabolomics #dataanalysis #zmnh #ukeHamburg
Now out in Acta Neuropathol Commun: actaneurocomms.biomedcentral.com/articles/10....
Special thanks to the Roggenbuck Foundation for funding!
#Neurooncology #Neuropathology #zmnh #ukeHamburg
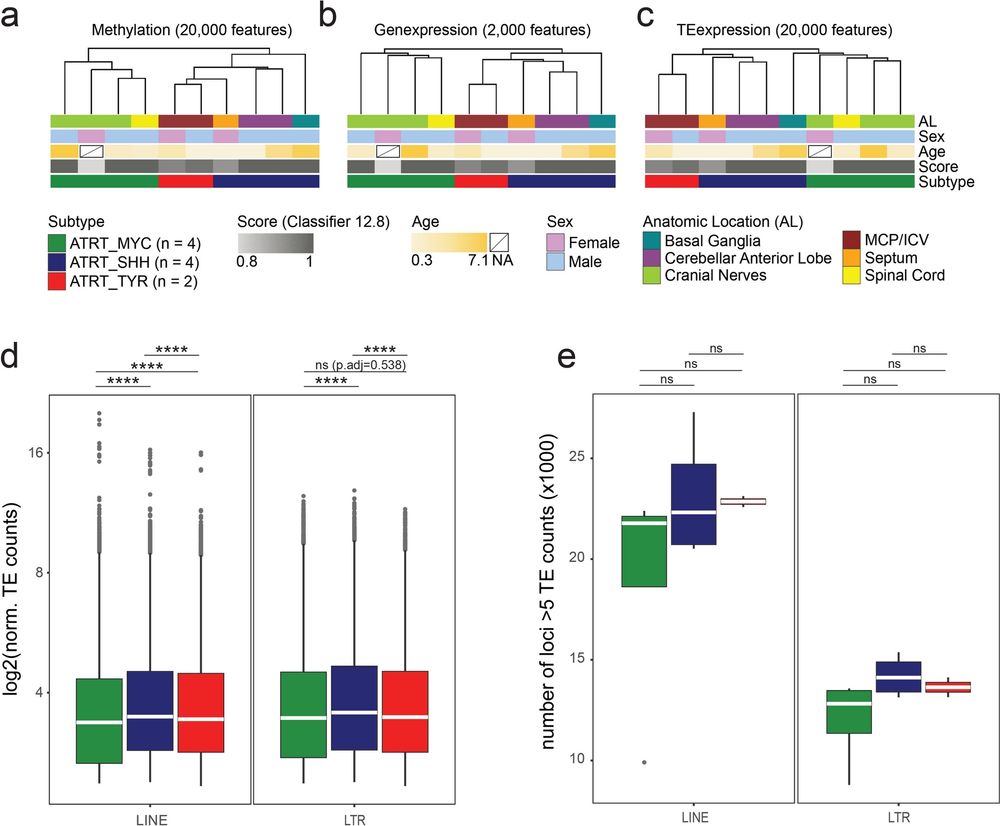
Now out in Acta Neuropathol Commun: actaneurocomms.biomedcentral.com/articles/10....
Special thanks to the Roggenbuck Foundation for funding!
#Neurooncology #Neuropathology #zmnh #ukeHamburg
#neurooncology #neurodevelopment #proteomics #Neuropathology #zmnh #ukeHamburg
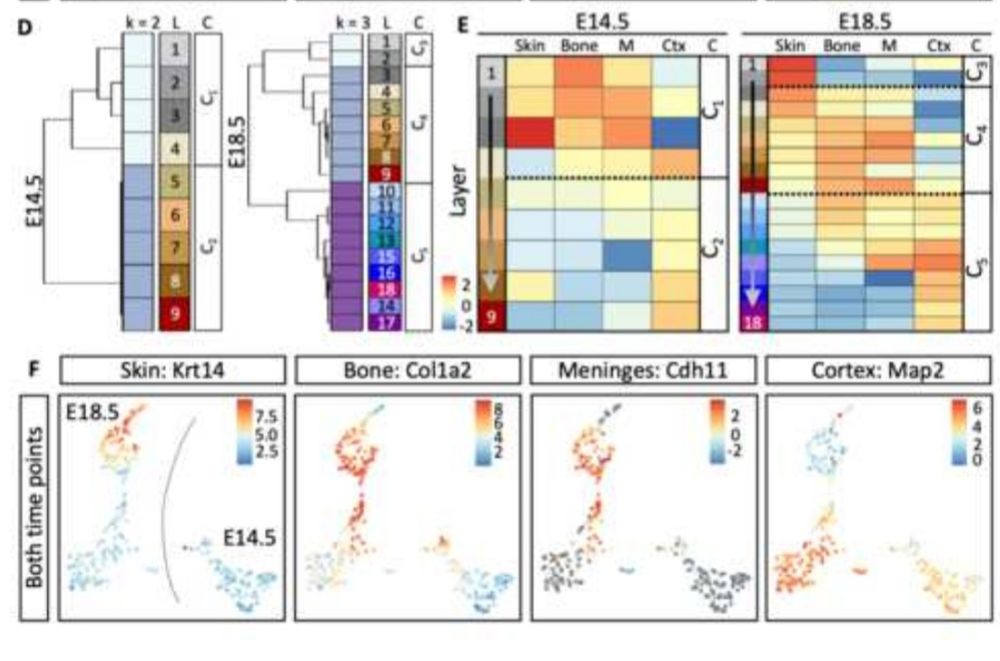
#neurooncology #neurodevelopment #proteomics #Neuropathology #zmnh #ukeHamburg
Read about outcome-associated factors in a molecularly defined cohort of central neurocytoma
in Acta Neuropathologica:
link.springer.com/article/10.1...
Happy to have contributed to the study!
#neurooncology #Neuropathology
#braintumor
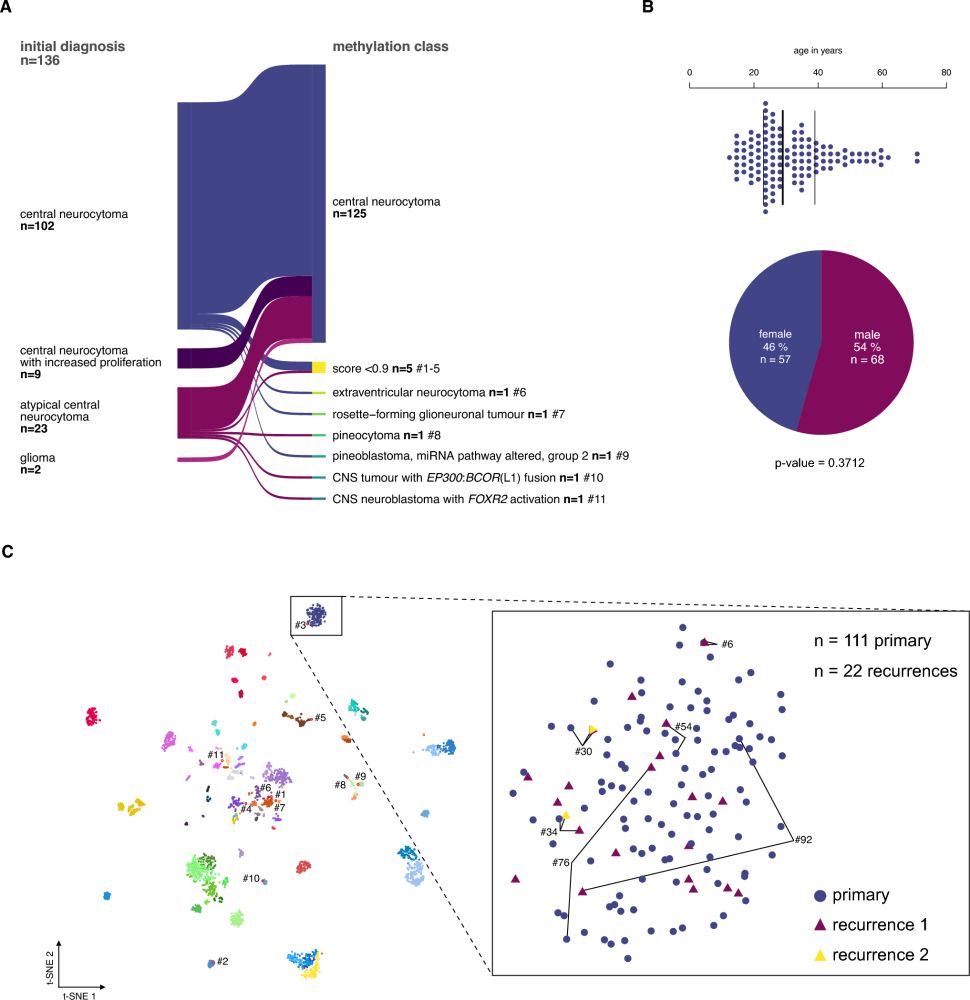
Read about outcome-associated factors in a molecularly defined cohort of central neurocytoma
in Acta Neuropathologica:
link.springer.com/article/10.1...
Happy to have contributed to the study!
#neurooncology #Neuropathology
#braintumor
She shows our data on LIN28A in atypical teratoid rhabdoid tumors and proteome data on a large cohort of #ependymoma. Fantastic job Antonia!
#atrt #braintumor #pediatriccancer #kinderkrebs #proteomics #neurooncology #Neuropathology #zmnh
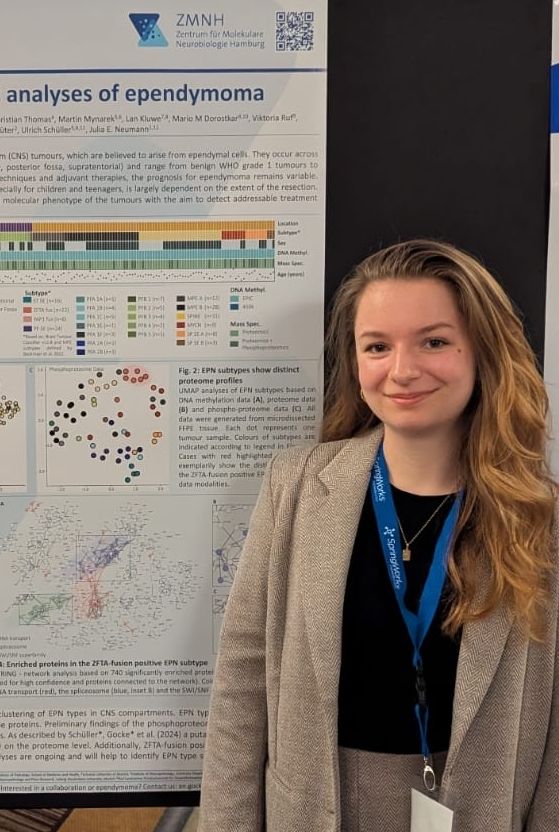
She shows our data on LIN28A in atypical teratoid rhabdoid tumors and proteome data on a large cohort of #ependymoma. Fantastic job Antonia!
#atrt #braintumor #pediatriccancer #kinderkrebs #proteomics #neurooncology #Neuropathology #zmnh
#proteomics #etmr #spatialproteomics #zmnh #ukeHamburg #neurooncology #neurodevelopment #neuropathology


#proteomics #etmr #spatialproteomics #zmnh #ukeHamburg #neurooncology #neurodevelopment #neuropathology
Have a look at our #review paper describing challenges of #data integration and available #algorithms and tools.
doi.org/10.1002/pmic...

Have a look at our #review paper describing challenges of #data integration and available #algorithms and tools.
doi.org/10.1002/pmic...
We provide significant improvements to the #HarmonizR approach:
1) Improved runtime
2) Retainment of more features (feature rescue of up to 103.9% for tested datasets)
link.springer.com/article/10.1...
#transcriptomics #proteomics #bioinformatics

We provide significant improvements to the #HarmonizR approach:
1) Improved runtime
2) Retainment of more features (feature rescue of up to 103.9% for tested datasets)
link.springer.com/article/10.1...
#transcriptomics #proteomics #bioinformatics

