
Computational biology | RNA Biology | RNA-binding proteins | Splicing | Aging | 3'UTR processing | lncRNA
https://profiles.imperial.ac.uk/n.haberman

We wrapped up insightful sessions on:
- RNA therapeutics & RNA modification
- RNA molecular machines & RNA-binding proteins
- RNA localization & turnover
Thanks to all speakers! @biochemsoc.bsky.social #BiochemEvent

We wrapped up insightful sessions on:
- RNA therapeutics & RNA modification
- RNA molecular machines & RNA-binding proteins
- RNA localization & turnover
Thanks to all speakers! @biochemsoc.bsky.social #BiochemEvent


In summary, we demonstrate that that capped 3′UTR-derived RNAs arise from structured G-motif-rich regions bound by AGO2 and UPF1, and they localise separately from their parental mRNAs.

In summary, we demonstrate that that capped 3′UTR-derived RNAs arise from structured G-motif-rich regions bound by AGO2 and UPF1, and they localise separately from their parental mRNAs.
Using high-resolution imaging, we validated that 3′UTR-derived RNAs localise differently from their parental mRNAs within the cell, hinting at potential independent functions.

Using high-resolution imaging, we validated that 3′UTR-derived RNAs localise differently from their parental mRNAs within the cell, hinting at potential independent functions.
We also found that cytoplasmic capping can occur subsequent to AGO2/siRNA-mediated cleavage, by analysing FANTOM5 CAGE from siRNA-treated samples. Our findings demonstrate that capping can readily follow cytoplasmic cleavage, and is not exclusive to transcriptional initiation.
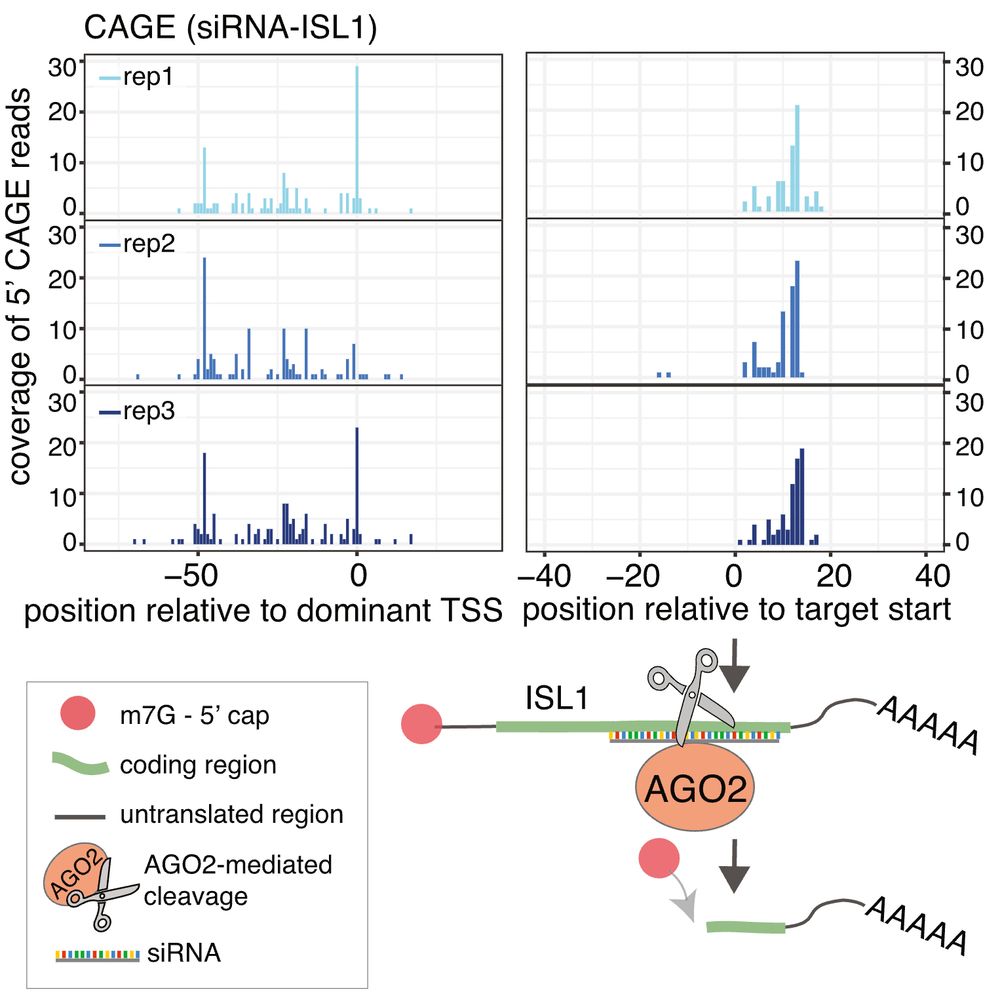
We also found that cytoplasmic capping can occur subsequent to AGO2/siRNA-mediated cleavage, by analysing FANTOM5 CAGE from siRNA-treated samples. Our findings demonstrate that capping can readily follow cytoplasmic cleavage, and is not exclusive to transcriptional initiation.
Our analysis shows that AGO2 binds upstream of 3′UTR-derived RNA start sites—independent of miRNA binding. Interestingly, G4 motifs influence the binding positions of both AGO2 and UPF1, shaping the capping sites.
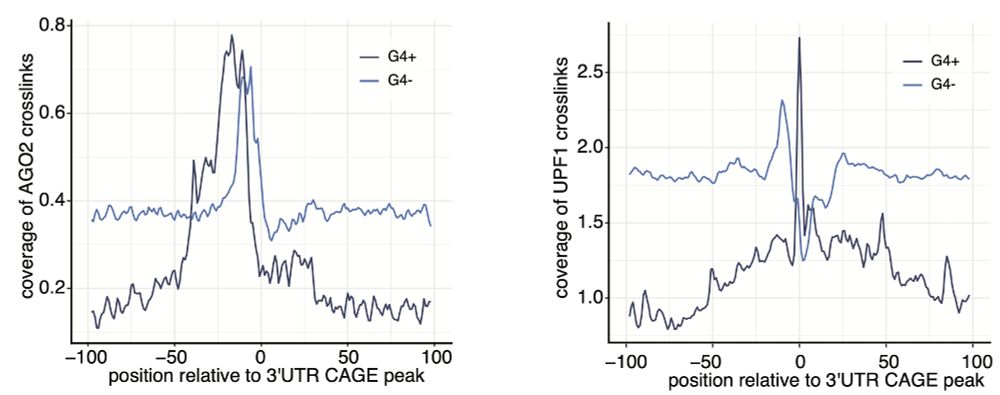
Our analysis shows that AGO2 binds upstream of 3′UTR-derived RNA start sites—independent of miRNA binding. Interestingly, G4 motifs influence the binding positions of both AGO2 and UPF1, shaping the capping sites.
By integrating AGO2-eiCLIP, eCLIP, and CAGE-seq, we found their origins linked to RNA-binding proteins, specifically AGO2 and UPF1. These sites are often rich in G-motifs capable of forming G-Quadruplexes.
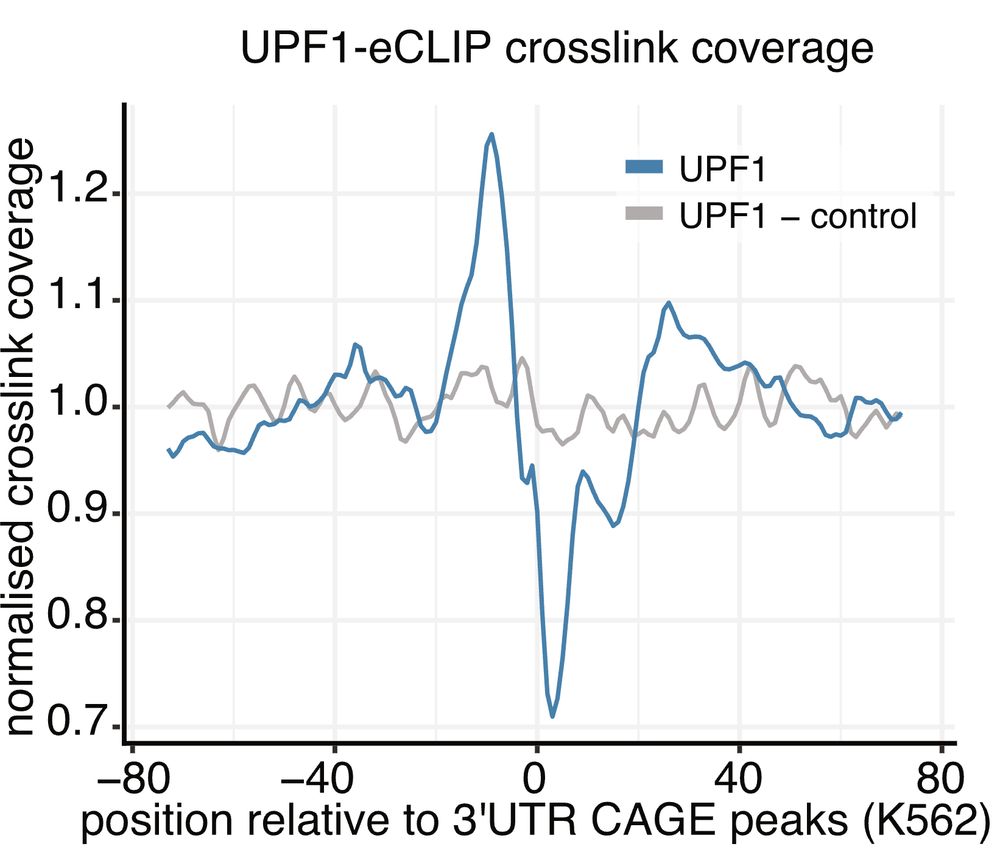

By integrating AGO2-eiCLIP, eCLIP, and CAGE-seq, we found their origins linked to RNA-binding proteins, specifically AGO2 and UPF1. These sites are often rich in G-motifs capable of forming G-Quadruplexes.
CAGE profiling often reveals signals (~10-15%) in 3′UTRs — regions far from usual transcription start sites (TSS). This phenomenon is reproducible but poorly understood.
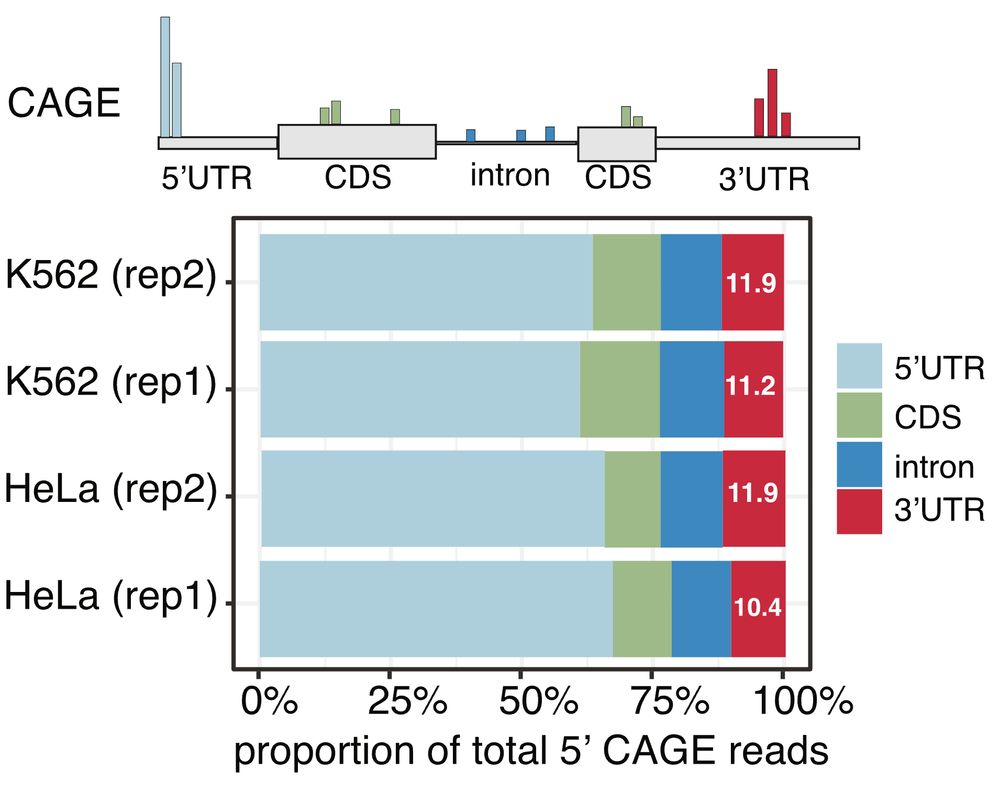
CAGE profiling often reveals signals (~10-15%) in 3′UTRs — regions far from usual transcription start sites (TSS). This phenomenon is reproducible but poorly understood.


