
More in our preprint: www.biorxiv.org/content/10.1...

More in our preprint: www.biorxiv.org/content/10.1...
Elyas Heidari and colleagues from the Gerstung, Pe'er and Stegle Labs released Segger, a new algorithm that uses GNN to model both transcripts and cells. More details in their preprint: www.biorxiv.org/content/10.1...

Elyas Heidari and colleagues from the Gerstung, Pe'er and Stegle Labs released Segger, a new algorithm that uses GNN to model both transcripts and cells. More details in their preprint: www.biorxiv.org/content/10.1...
Our new study on Xenium exploration best practice and comparing it with other commercial platforms is now out in @naturemethods.bsky.social
www.nature.com/articles/s41...
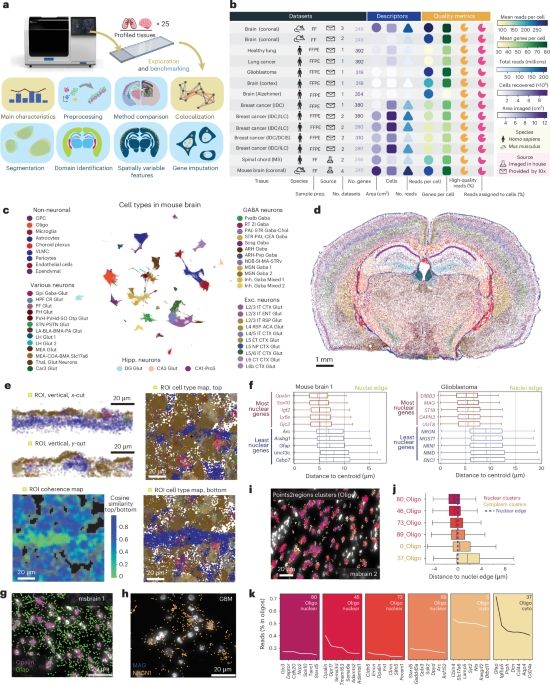
Our new study on Xenium exploration best practice and comparing it with other commercial platforms is now out in @naturemethods.bsky.social
www.nature.com/articles/s41...

Spread the word!

Spread the word!

More here: spatialhackathon.github.io

More here: spatialhackathon.github.io
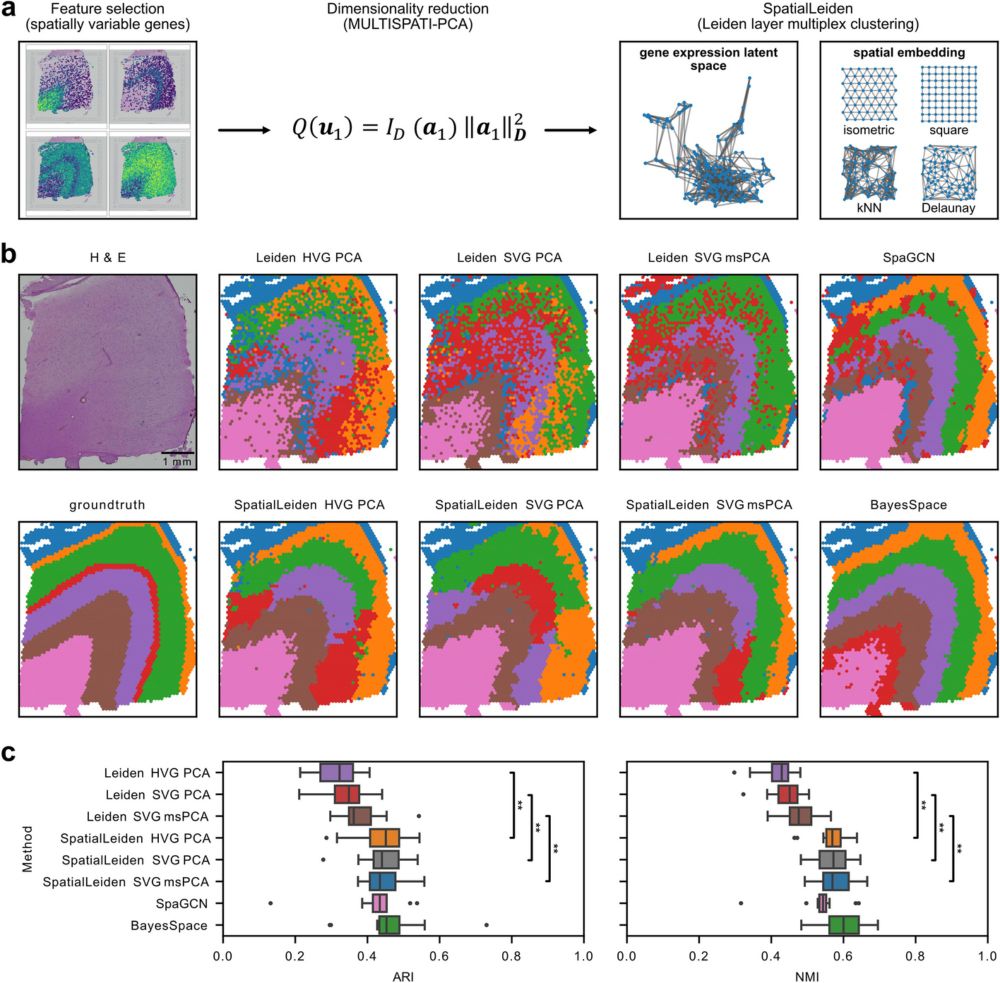
github.com/HiDiHlabs/sp...
It's as easy as Leiden, but as performant as the best in class. Give it a try and let us know if it works for you!

github.com/HiDiHlabs/sp...
It's as easy as Leiden, but as performant as the best in class. Give it a try and let us know if it works for you!
#Sainsc can map cell types in high resolution spatial transcriptomics data, without segmenting cells! It's light weight, fast, and easy to use: sainsc.readthedocs.io/tutorials/in...

#Sainsc can map cell types in high resolution spatial transcriptomics data, without segmenting cells! It's light weight, fast, and easy to use: sainsc.readthedocs.io/tutorials/in...
Incredible study - 3D single cell imaging of the entire mouse brain - also on the cover of this week's #Science
🧪🔬🤯
www.science.org/doi/10.1126/...

Incredible study - 3D single cell imaging of the entire mouse brain - also on the cover of this week's #Science
🧪🔬🤯
www.science.org/doi/10.1126/...

