Nathan Grubaugh
@nathangrubaugh.bsky.social
Associate Professor YaleEMD / YaleSPH. Studies 🦟🦠 🧬transmission, evolution, and emergence. PI of http://dengue-lineages.org. Will sequence for 🍺.
Lab website: https://grubaughlab.com/
Lab website: https://grubaughlab.com/
Along the way, we discuss a few important topics:
- Approaches to sequencing arboviruses (Box 1)
- Discrete phylogenetic approaches for determining arbovirus transmission histories (Box 2, 👇 )
- How to deal with sampling biases (main text)
- Future directions of using phylogenetics (conclusions)
- Approaches to sequencing arboviruses (Box 1)
- Discrete phylogenetic approaches for determining arbovirus transmission histories (Box 2, 👇 )
- How to deal with sampling biases (main text)
- Future directions of using phylogenetics (conclusions)

June 9, 2025 at 2:03 PM
Along the way, we discuss a few important topics:
- Approaches to sequencing arboviruses (Box 1)
- Discrete phylogenetic approaches for determining arbovirus transmission histories (Box 2, 👇 )
- How to deal with sampling biases (main text)
- Future directions of using phylogenetics (conclusions)
- Approaches to sequencing arboviruses (Box 1)
- Discrete phylogenetic approaches for determining arbovirus transmission histories (Box 2, 👇 )
- How to deal with sampling biases (main text)
- Future directions of using phylogenetics (conclusions)
Finally, we discuss integrating ecological and epidemiological covariate data into the phylodynamic framework to discover the complex drivers of transmission and spread.

June 9, 2025 at 2:03 PM
Finally, we discuss integrating ecological and epidemiological covariate data into the phylodynamic framework to discover the complex drivers of transmission and spread.
We then detail how using phylodynamic tools, including phylogeography, can reveal detailed patterns of spillovers from sylvatic cycles (e.g. below) and invasion into new regions.

June 9, 2025 at 2:03 PM
We then detail how using phylodynamic tools, including phylogeography, can reveal detailed patterns of spillovers from sylvatic cycles (e.g. below) and invasion into new regions.
We highlight how arbovirus genetic diversity can better inform classification & within-species dynamics and how some simple phylogenetic methods can help to answer important questions about virus emergence and spread.

June 9, 2025 at 2:03 PM
We highlight how arbovirus genetic diversity can better inform classification & within-species dynamics and how some simple phylogenetic methods can help to answer important questions about virus emergence and spread.
Arboviruses are highly diverse in terms of virus families / genome structures and types of vectors and hosts. We tried to draw upon as many different examples as possible and learned a ton about some less studied viruses along the way! (e.g. Toscana and bluetongue are some of my new favs).

June 9, 2025 at 2:02 PM
Arboviruses are highly diverse in terms of virus families / genome structures and types of vectors and hosts. We tried to draw upon as many different examples as possible and learned a ton about some less studied viruses along the way! (e.g. Toscana and bluetongue are some of my new favs).
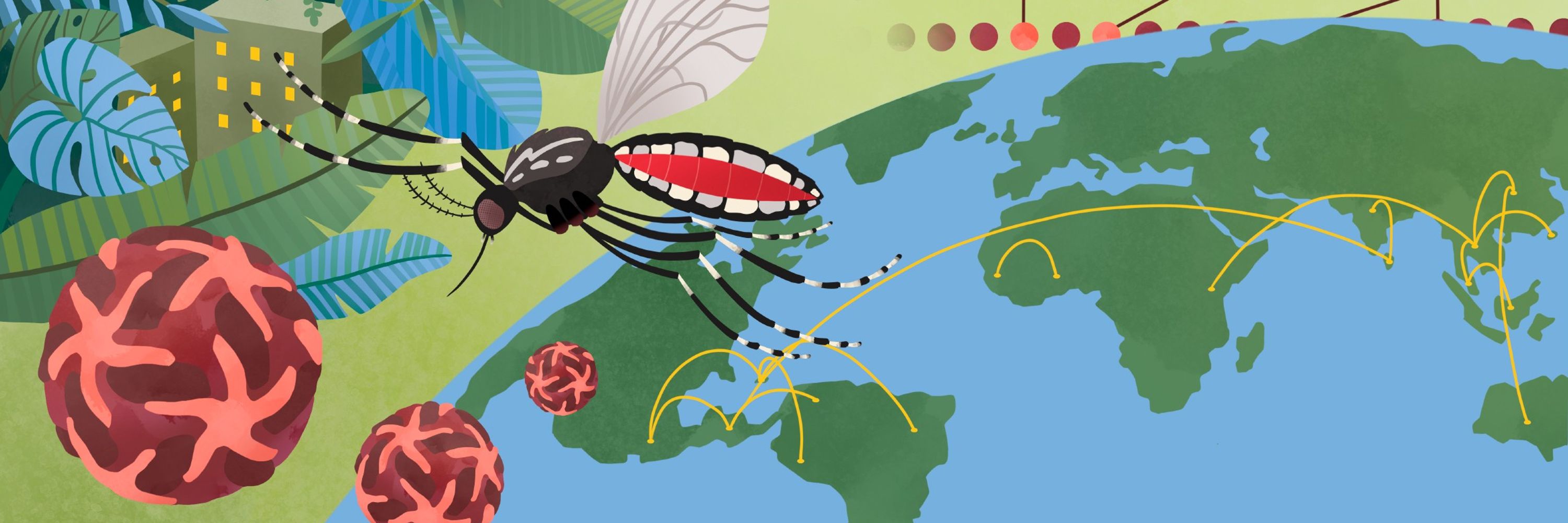
Interested in using phylogenetics to study arboviruses? Our new review in @natrevgenet.nature.com by @viralverity.bsky.social, @sdellicour.bsky.social, and Marta Giovanetti has you covered!
📖 👉 rdcu.be/epH2U
Short 🧵
📖 👉 rdcu.be/epH2U
Short 🧵

June 9, 2025 at 2:02 PM
Interested in using phylogenetics to study arboviruses? Our new review in @natrevgenet.nature.com by @viralverity.bsky.social, @sdellicour.bsky.social, and Marta Giovanetti has you covered!
📖 👉 rdcu.be/epH2U
Short 🧵
📖 👉 rdcu.be/epH2U
Short 🧵
What a sequence

March 28, 2025 at 2:45 AM
What a sequence
Sharing our work expanding SARS-CoV-2 style tiled amplicon sequencing for whole bacteria genomes directly from clinical samples, showing examples with M.tb (>4 mb) and Strep pneumo (>2 mb).
Led by Chaney Kalinich, Freddy Gonzalez, & Seth Redmond
www.biorxiv.org/content/10.1...
Led by Chaney Kalinich, Freddy Gonzalez, & Seth Redmond
www.biorxiv.org/content/10.1...

December 20, 2024 at 8:45 PM
Sharing our work expanding SARS-CoV-2 style tiled amplicon sequencing for whole bacteria genomes directly from clinical samples, showing examples with M.tb (>4 mb) and Strep pneumo (>2 mb).
Led by Chaney Kalinich, Freddy Gonzalez, & Seth Redmond
www.biorxiv.org/content/10.1...
Led by Chaney Kalinich, Freddy Gonzalez, & Seth Redmond
www.biorxiv.org/content/10.1...
Unfortunately, this is my house right now

December 20, 2024 at 12:09 AM
Unfortunately, this is my house right now
For transparency - here is everything that I said to @cofford.bsky.social - who wrote this very good article.
While i think that the names are mostly "silly and pompous", I don't think that this will really have much impact either way. Annoying, but meh.
While i think that the names are mostly "silly and pompous", I don't think that this will really have much impact either way. Annoying, but meh.

December 13, 2024 at 9:14 PM
For transparency - here is everything that I said to @cofford.bsky.social - who wrote this very good article.
While i think that the names are mostly "silly and pompous", I don't think that this will really have much impact either way. Annoying, but meh.
While i think that the names are mostly "silly and pompous", I don't think that this will really have much impact either way. Annoying, but meh.
Lab decorated the lab with sleigh of Handsome Dans 🎅


December 4, 2024 at 6:56 PM
Lab decorated the lab with sleigh of Handsome Dans 🎅
Pre-dinner cocktails: cranberry aperol spritz and cider mimosas.
After dinner: Apple pie old fashioneds using the infused bourbon in the jar.
Cheers!
After dinner: Apple pie old fashioneds using the infused bourbon in the jar.
Cheers!

November 28, 2024 at 7:17 PM
Pre-dinner cocktails: cranberry aperol spritz and cider mimosas.
After dinner: Apple pie old fashioneds using the infused bourbon in the jar.
Cheers!
After dinner: Apple pie old fashioneds using the infused bourbon in the jar.
Cheers!
Drop us a line if you have any questions, suggestions - or just want to collaborate.
dengue-lineages.org/contact.html
dengue-lineages.org/contact.html

November 18, 2024 at 10:53 PM
Drop us a line if you have any questions, suggestions - or just want to collaborate.
dengue-lineages.org/contact.html
dengue-lineages.org/contact.html
We also have several other resources:
Designate a new lineage: dengue-lineages.org/design.html
Track existing lineages: dengue-lineages.org/lineages.html
Genome surveillance resources: dengue-lineages.org/resources.html
Designate a new lineage: dengue-lineages.org/design.html
Track existing lineages: dengue-lineages.org/lineages.html
Genome surveillance resources: dengue-lineages.org/resources.html

November 18, 2024 at 10:53 PM
We also have several other resources:
Designate a new lineage: dengue-lineages.org/design.html
Track existing lineages: dengue-lineages.org/lineages.html
Genome surveillance resources: dengue-lineages.org/resources.html
Designate a new lineage: dengue-lineages.org/design.html
Track existing lineages: dengue-lineages.org/lineages.html
Genome surveillance resources: dengue-lineages.org/resources.html
You can assign lineages to your sequences using several tools that are available on our website.

November 18, 2024 at 10:53 PM
You can assign lineages to your sequences using several tools that are available on our website.
Our system splits previously defined dengue virus serotypes and genotypes into major and minor lineages to better enable tracking and characterization of genetic diversity.

November 18, 2024 at 10:53 PM
Our system splits previously defined dengue virus serotypes and genotypes into major and minor lineages to better enable tracking and characterization of genetic diversity.
If you do dengue virus sequencing +/- phylogenetic analysis, check out our new system for classifying lineages.
Led by Verity Hill, this was a large collaborative effort by dengue researchers from 14 countries.
Paper -> journals.plos.org/plosbiology/...
Website-> dengue-lineages.org
Led by Verity Hill, this was a large collaborative effort by dengue researchers from 14 countries.
Paper -> journals.plos.org/plosbiology/...
Website-> dengue-lineages.org


November 18, 2024 at 9:58 PM
If you do dengue virus sequencing +/- phylogenetic analysis, check out our new system for classifying lineages.
Led by Verity Hill, this was a large collaborative effort by dengue researchers from 14 countries.
Paper -> journals.plos.org/plosbiology/...
Website-> dengue-lineages.org
Led by Verity Hill, this was a large collaborative effort by dengue researchers from 14 countries.
Paper -> journals.plos.org/plosbiology/...
Website-> dengue-lineages.org


