


Joint project with Gregoire Montavon’s lab at #BIFOLD @xtraexer.bsky.social -
explainable AI for self supervised LLMs in gene regulation. Apply until Dec 13!
www.bifold.berlin/about-us/opp...
Joint project with Gregoire Montavon’s lab at #BIFOLD @xtraexer.bsky.social -
explainable AI for self supervised LLMs in gene regulation. Apply until Dec 13!
www.bifold.berlin/about-us/opp...
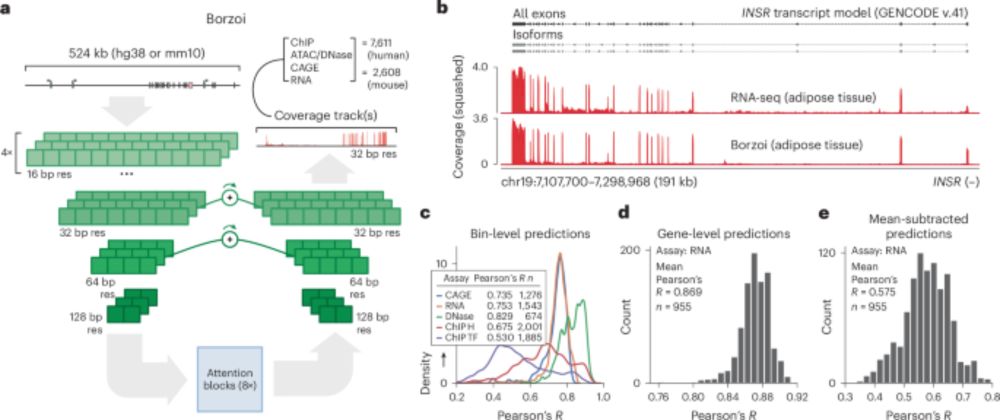
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Please Repost to spread the word! 🙏🏻
Led by @zrcjessica
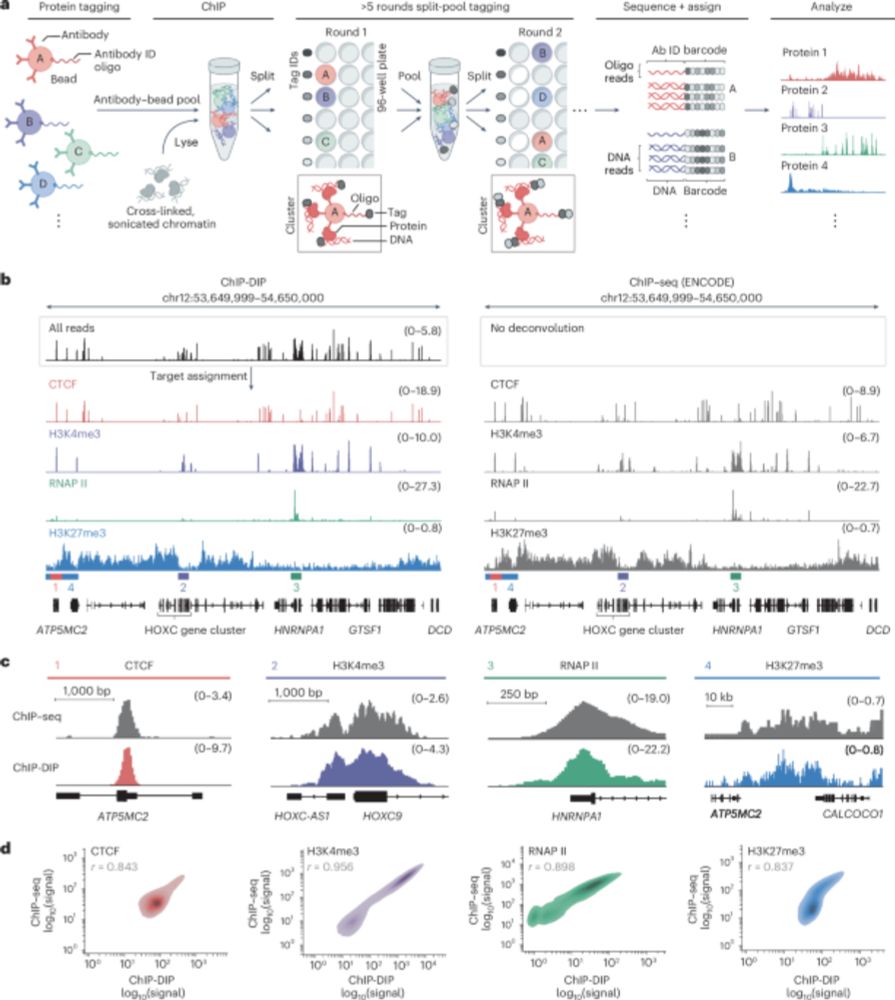
Paper: www.biorxiv.org/content/10.1...
2/n

Please Repost to spread the word! 🙏🏻

