github.com/quevan/phlame
github.com/quevan/phlame
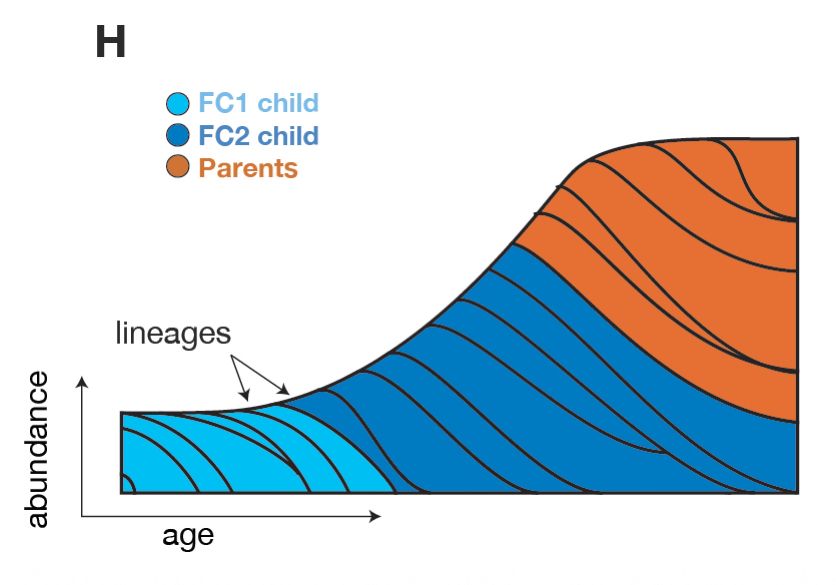
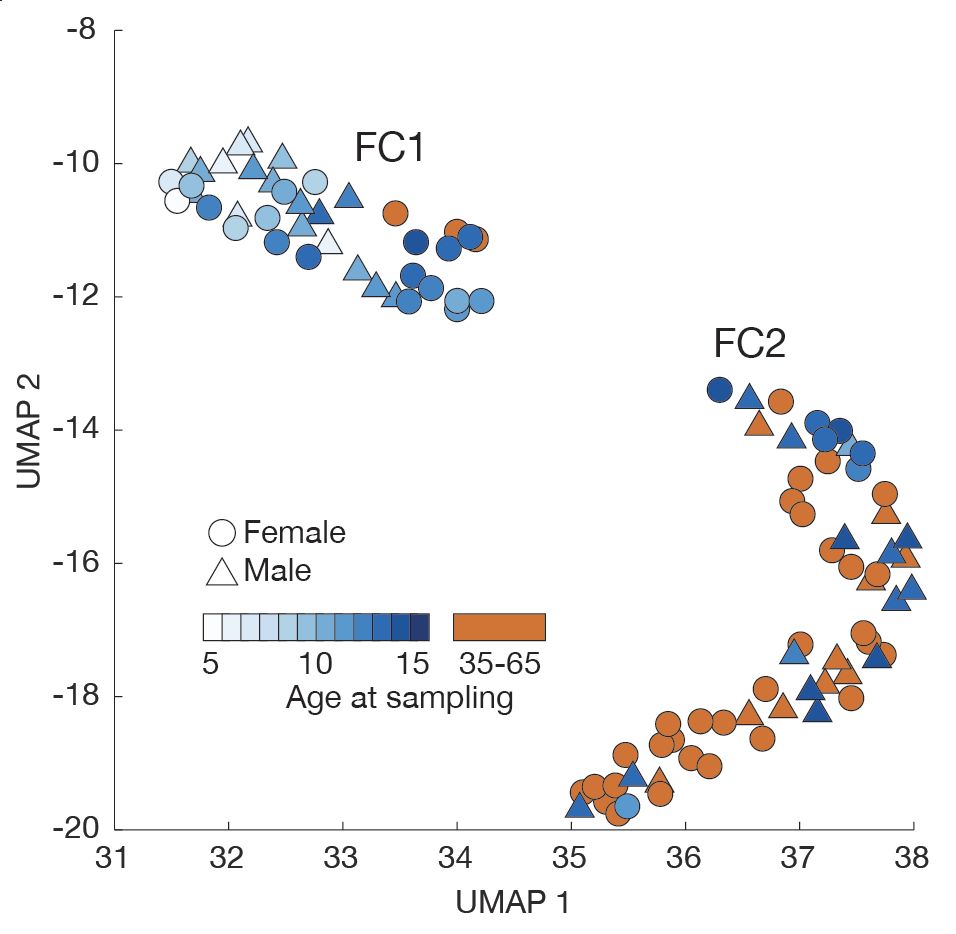
📈🦠 We find that C. acnes colonization surges in adolescence during population expansion, apparently alleviating barriers to colonization before leveling off in adulthood. This period seems crucial for shaping the skin microbiome long-term.
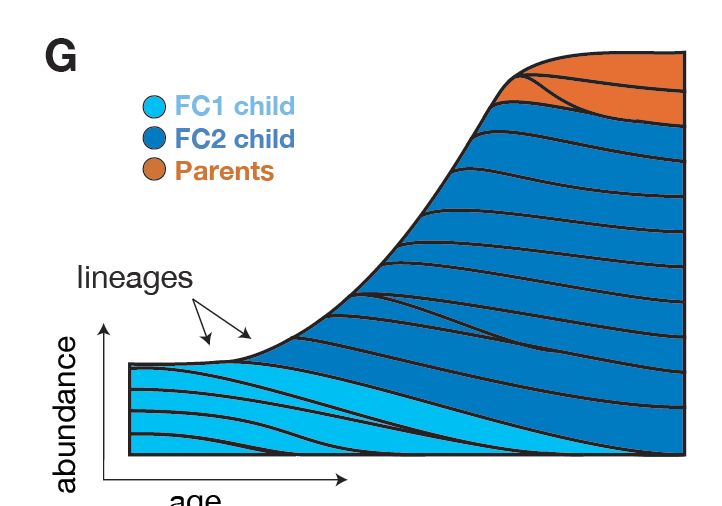
📈🦠 We find that C. acnes colonization surges in adolescence during population expansion, apparently alleviating barriers to colonization before leveling off in adulthood. This period seems crucial for shaping the skin microbiome long-term.