
Michael Glickman
@michaelglickman.bsky.social
Physician Scientist interested in all things mycobacterial: pathogen, model system, and cancer therapy. Immunology program/ID MSK. Opinions are mine and do not represent those of my employer, MSK.
https://scholar.google.com/citations?hl=en&user=xriihOcAA
https://scholar.google.com/citations?hl=en&user=xriihOcAA
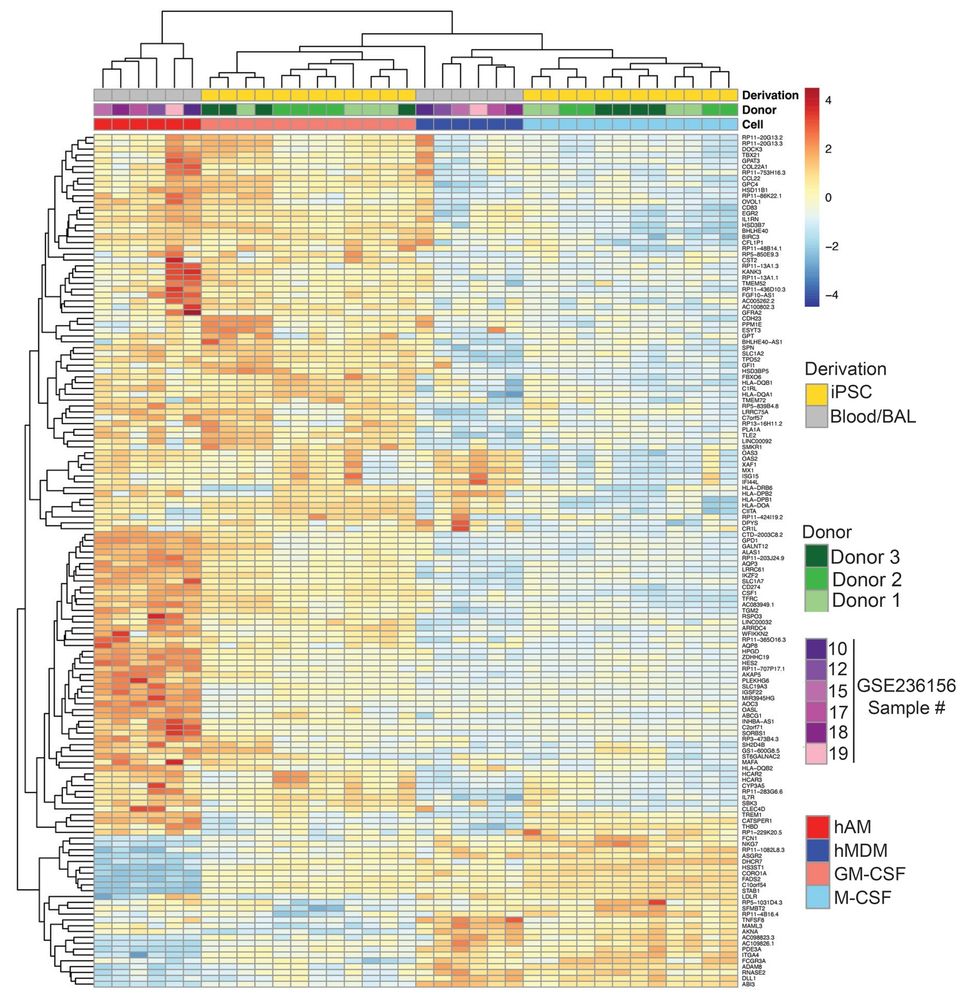
We show that these iPSC derived alveolar macrophages, which we call AM-L (like), resemble bonafide AMs at baseline and with Mtb infection, leveraging the insights of our collaborators in the Hawn lab published here:
journals.aai.org/jimmunol/art...
journals.aai.org/jimmunol/art...
July 28, 2025 at 9:43 PM
We show that these iPSC derived alveolar macrophages, which we call AM-L (like), resemble bonafide AMs at baseline and with Mtb infection, leveraging the insights of our collaborators in the Hawn lab published here:
journals.aai.org/jimmunol/art...
journals.aai.org/jimmunol/art...
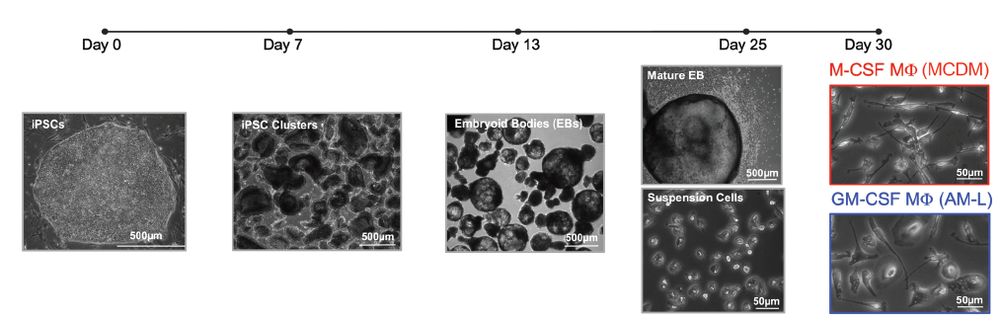
To faciliate human AM studies, we derived macrophage subtypes from iPSCs under distinct cytokines, including GM-CSF, which is required for development of AMs.
July 28, 2025 at 9:43 PM
To faciliate human AM studies, we derived macrophage subtypes from iPSCs under distinct cytokines, including GM-CSF, which is required for development of AMs.
And also show that placing the ALFA tag at different places in a multidomain protein (Pks13) allows for domain specific interactomes:

March 25, 2025 at 2:54 PM
And also show that placing the ALFA tag at different places in a multidomain protein (Pks13) allows for domain specific interactomes:
And can be used for proximity proteomics by fusing NBALFA to TurboID. We present a full workflow for proximity proteomics in M. smegmatis and M. tuberculosis. Here shown for the mycolic acid biosynthetic protein Pks13:

March 25, 2025 at 2:54 PM
And can be used for proximity proteomics by fusing NBALFA to TurboID. We present a full workflow for proximity proteomics in M. smegmatis and M. tuberculosis. Here shown for the mycolic acid biosynthetic protein Pks13:
Allison reasoned that placing the ALFA tag on the target protein and expressing the high affinity anti-ALFA nanobody fused to various functional domains in live mycobacteria would be a versatile system. We show that such an approach enables faitful localization of proteins, here shown for MmpL3...

March 25, 2025 at 2:54 PM
Allison reasoned that placing the ALFA tag on the target protein and expressing the high affinity anti-ALFA nanobody fused to various functional domains in live mycobacteria would be a versatile system. We show that such an approach enables faitful localization of proteins, here shown for MmpL3...
Important point, the name is misleading, as most names are. Here is the cdc map from 2019 for spotted fever group rickettsiosis.

December 9, 2023 at 5:22 PM
Important point, the name is misleading, as most names are. Here is the cdc map from 2019 for spotted fever group rickettsiosis.


