📍 MICROBIOME session
Weds evening | Room 01B
📍 MICROBIOME session
Weds evening | Room 01B
📍 MICROBIOME session
Weds evening | Room 01B
📍 MICROBIOME session
Weds evening | Room 01B
📍ELIXIR/NIH-ODSS session
Weds afternoon | Room 12
📍ELIXIR/NIH-ODSS session
Weds afternoon | Room 12
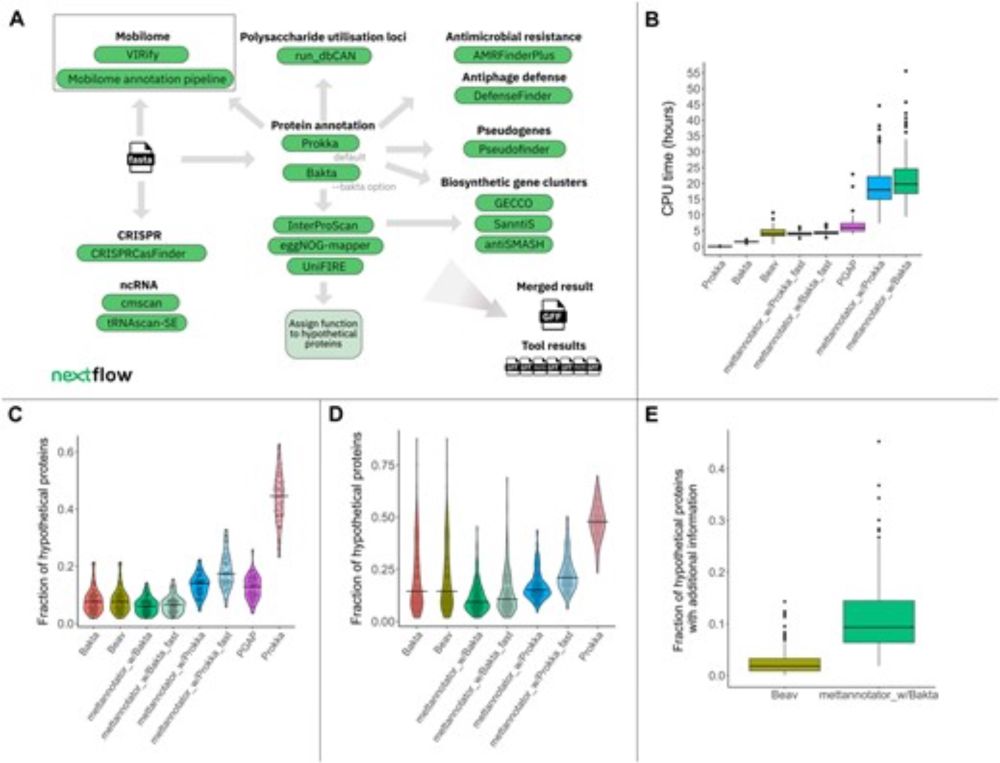
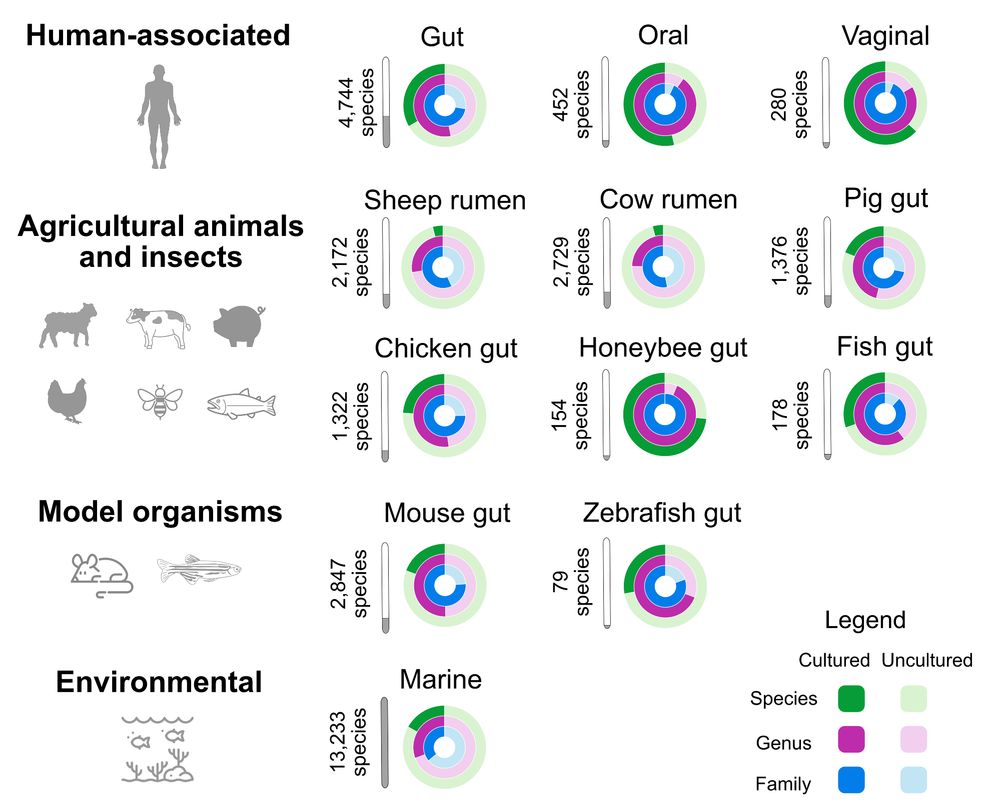
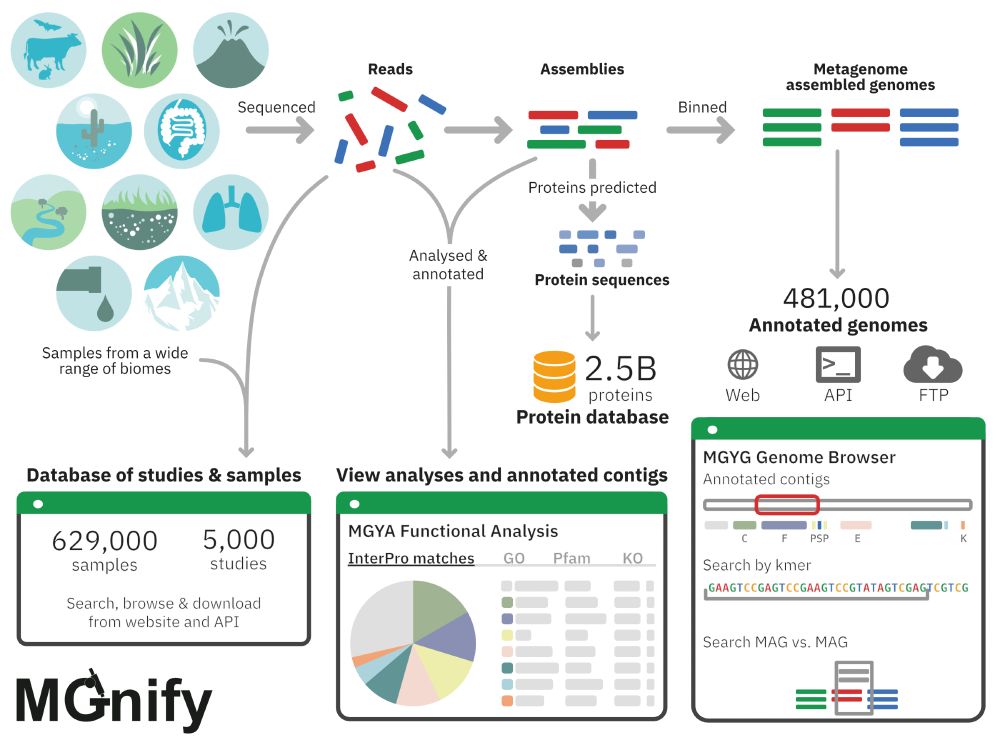

🔎Protein structures @pdbeurope.bsky.social
🌍Metagenomic data @Mgnifydb.bsky.social
🔎Protein families & functional sites @interprodb.bsky.social
🧪Bioactive molecules & drug discovery @chembl.bsky.social
And more!!
🔎Protein structures @pdbeurope.bsky.social
🌍Metagenomic data @Mgnifydb.bsky.social
🔎Protein families & functional sites @interprodb.bsky.social
🧪Bioactive molecules & drug discovery @chembl.bsky.social
And more!!

