

Mutations are quite biased towards G:C ➡️ A:T and A:T➡️G:C; and this is very apparent in L. acidophilus near the ori.

Mutations are quite biased towards G:C ➡️ A:T and A:T➡️G:C; and this is very apparent in L. acidophilus near the ori.

We are looking for a research assistant who will be involved with our experimental evolution studies on how stress affects population dynamics in E. coli.
Great opportunity for recent grads!
E-mail for more info and apply here:
ecsr.fa.us2.oraclecloud.com/hcmUI/Candid...

We are looking for a research assistant who will be involved with our experimental evolution studies on how stress affects population dynamics in E. coli.
Great opportunity for recent grads!
E-mail for more info and apply here:
ecsr.fa.us2.oraclecloud.com/hcmUI/Candid...



www.pnas.org/doi/full/10....

www.pnas.org/doi/full/10....
A great example of how lab evolution can provide insight into the natural world!
A great example of how lab evolution can provide insight into the natural world!

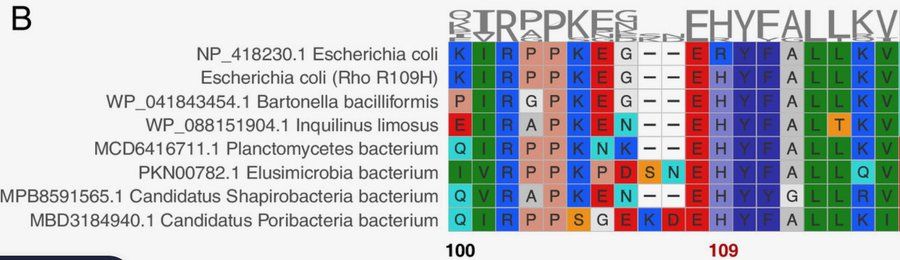
(These substitutions helped us get a better candidate YdcI structure)
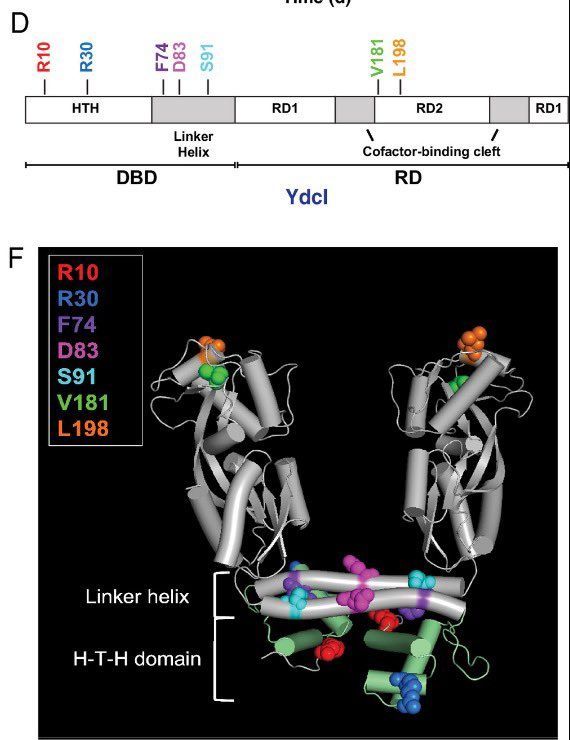
(These substitutions helped us get a better candidate YdcI structure)
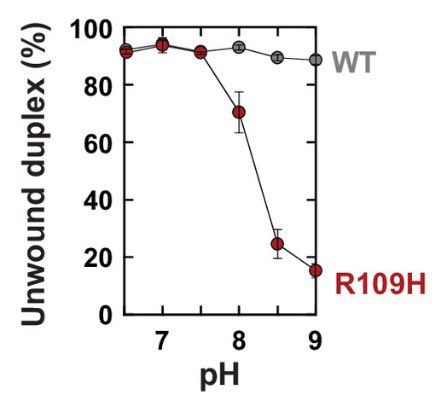
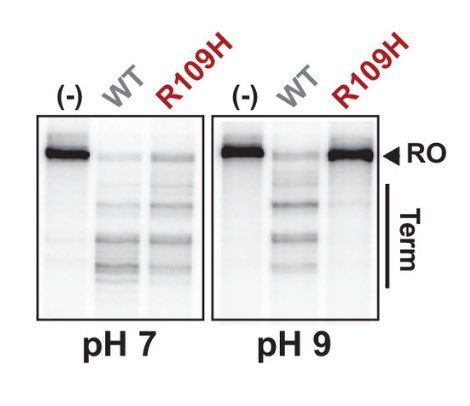
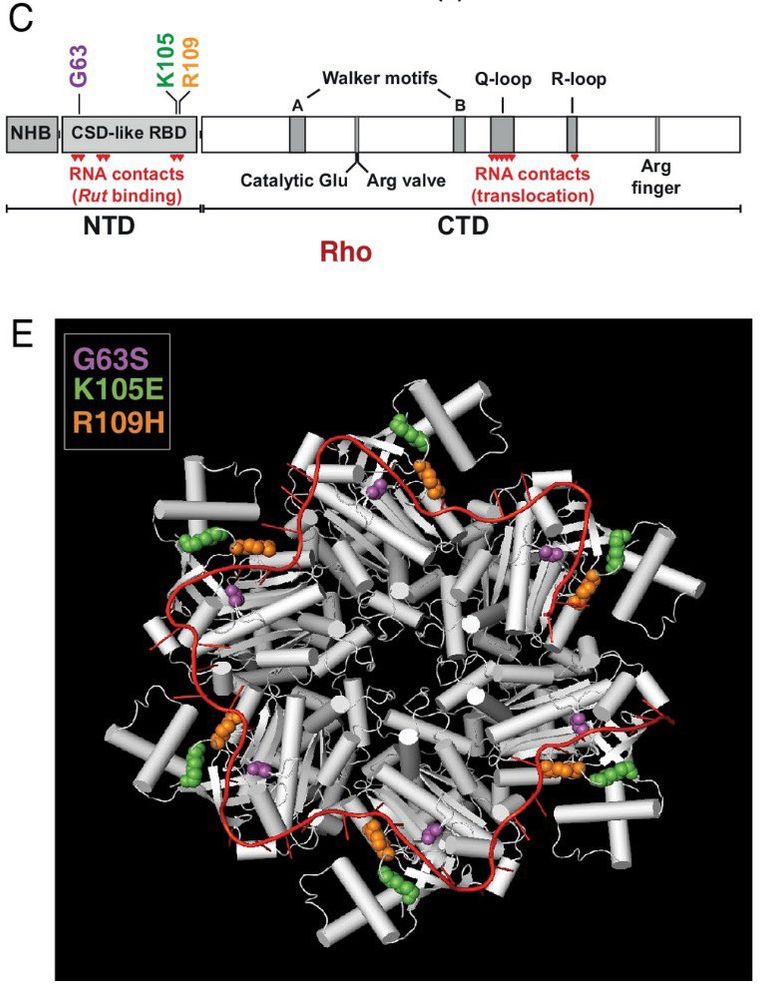
Our new paper led by postdoc @sb_worthan examining a pH sensitive substitution in the Rho transcription terminator that evolved experimentally in our lab and then is found in natural populations! Thread below! 🧵
www.pnas.org/doi/full/10....
Our new paper led by postdoc @sb_worthan examining a pH sensitive substitution in the Rho transcription terminator that evolved experimentally in our lab and then is found in natural populations! Thread below! 🧵
www.pnas.org/doi/full/10....
Thanks for letting this biologist join, there’s no better way to watch a game than in the VU box! ⚽️⚓️🧑🏫

Thanks for letting this biologist join, there’s no better way to watch a game than in the VU box! ⚽️⚓️🧑🏫
If you are at #TAGC24, Poster #777🎰 covers this & our follow up work on pH sensing mutations in transcription termination!

If you are at #TAGC24, Poster #777🎰 covers this & our follow up work on pH sensing mutations in transcription termination!
Exciting collaboration between my group, @MarcBoudvillain’s group and @bpbratton!
From populations experimentally evolved to extreme feast/famine cycles, we find pH-sensing rho alleles that confer pH-dependent plasticity in transcription termination!


Exciting collaboration between my group, @MarcBoudvillain’s group and @bpbratton!
From populations experimentally evolved to extreme feast/famine cycles, we find pH-sensing rho alleles that confer pH-dependent plasticity in transcription termination!
journals.asm.org/doi/10.1128/...



journals.asm.org/doi/10.1128/...

