MC Gambetta
@mcgambetta.bsky.social
Associate Professor at Center for Integrative Genomics (University of Lausanne)
This work was led by Marion Mouginot, who notably established a locus-specific proteomics approach that identified Lola-I as a meta-loop-associated factor,
together with Sahar Hani (supported by Arianna Ravera, UNIL’s image analysis expert!), who uncovered hierarchical meta-loop formation. 👏
together with Sahar Hani (supported by Arianna Ravera, UNIL’s image analysis expert!), who uncovered hierarchical meta-loop formation. 👏
April 18, 2025 at 6:50 PM
This work was led by Marion Mouginot, who notably established a locus-specific proteomics approach that identified Lola-I as a meta-loop-associated factor,
together with Sahar Hani (supported by Arianna Ravera, UNIL’s image analysis expert!), who uncovered hierarchical meta-loop formation. 👏
together with Sahar Hani (supported by Arianna Ravera, UNIL’s image analysis expert!), who uncovered hierarchical meta-loop formation. 👏
Lola-I was, however, only required to form a single meta-domain. This highlights a dichotomy between factors like Lola-I, which may control neuronal fate largely independently of inter-TAD interactions, and structural proteins like Cp190, which may enable ultra-long range gene regulation.

April 18, 2025 at 6:50 PM
Lola-I was, however, only required to form a single meta-domain. This highlights a dichotomy between factors like Lola-I, which may control neuronal fate largely independently of inter-TAD interactions, and structural proteins like Cp190, which may enable ultra-long range gene regulation.
We developed an unbiased proteomics approach (inspired by Baldi..Becker 2018) to identify proteins bound to meta-loop anchors. This identified a specific isoform of the axon guidance protein Longitudinals lacking (Lola-I) as enriched at several meta-loop anchors, in our pull-downs and in vivo.

April 18, 2025 at 6:50 PM
We developed an unbiased proteomics approach (inspired by Baldi..Becker 2018) to identify proteins bound to meta-loop anchors. This identified a specific isoform of the axon guidance protein Longitudinals lacking (Lola-I) as enriched at several meta-loop anchors, in our pull-downs and in vivo.
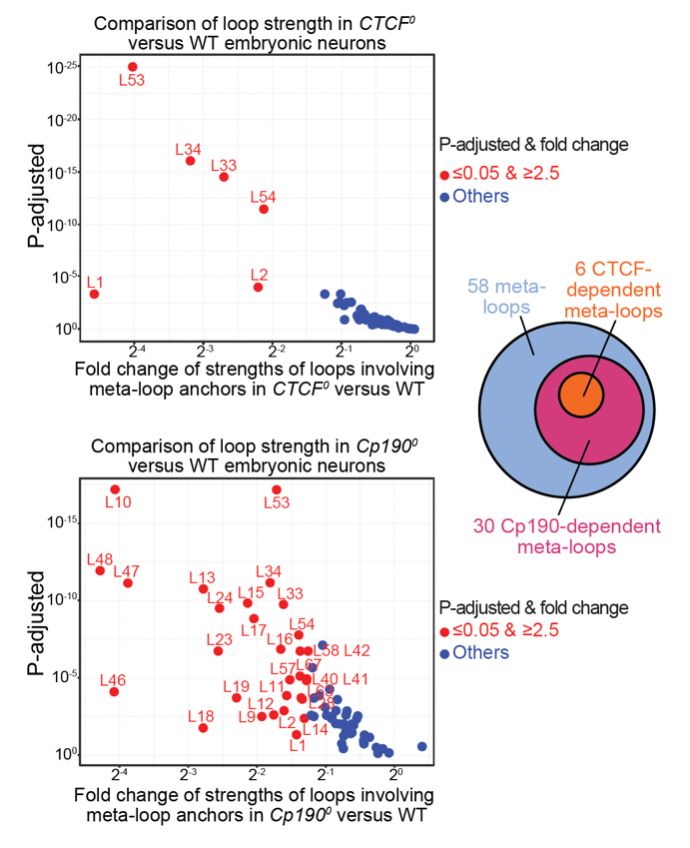
Many meta-loop anchors coincide with TAD boundaries. We found that a major determinant of TAD boundary formation in flies (called Cp190) is required to form a large fraction of meta-loops, including CTCF-dependent meta-loops.
April 18, 2025 at 6:50 PM
Many meta-loop anchors coincide with TAD boundaries. We found that a major determinant of TAD boundary formation in flies (called Cp190) is required to form a large fraction of meta-loops, including CTCF-dependent meta-loops.
How do meta-loops form?
Most meta-domains contain 2 or more meta-loops, with some of these forming earlier during neurogenesis than the other(s) in the same meta-domain.
We found that in many meta-domains, specific meta-loops (often the earlier forming ones) nucleate others.
Most meta-domains contain 2 or more meta-loops, with some of these forming earlier during neurogenesis than the other(s) in the same meta-domain.
We found that in many meta-domains, specific meta-loops (often the earlier forming ones) nucleate others.

April 18, 2025 at 6:50 PM
How do meta-loops form?
Most meta-domains contain 2 or more meta-loops, with some of these forming earlier during neurogenesis than the other(s) in the same meta-domain.
We found that in many meta-domains, specific meta-loops (often the earlier forming ones) nucleate others.
Most meta-domains contain 2 or more meta-loops, with some of these forming earlier during neurogenesis than the other(s) in the same meta-domain.
We found that in many meta-domains, specific meta-loops (often the earlier forming ones) nucleate others.
1 meta-domain we studied contains a meta-loop connecting promoters of Glutamate receptors 1A and 1B. Disrupting the meta-domain uncoupled their co-transcription (similar to Levo..Levine 2022 for intra-TAD loops connecting paralogous gene promoters).

April 18, 2025 at 6:50 PM
1 meta-domain we studied contains a meta-loop connecting promoters of Glutamate receptors 1A and 1B. Disrupting the meta-domain uncoupled their co-transcription (similar to Levo..Levine 2022 for intra-TAD loops connecting paralogous gene promoters).
We precisely disrupted 3 independent meta-domains and found transcriptional defects in all 3 cases.
2 meta-domains we studied contain a meta-loop connecting a neuronal gene promoter to a distant intergenic element. Disrupting the meta-domain decreased neuronal gene transcription by 2-to-3-fold.
2 meta-domains we studied contain a meta-loop connecting a neuronal gene promoter to a distant intergenic element. Disrupting the meta-domain decreased neuronal gene transcription by 2-to-3-fold.
April 18, 2025 at 6:50 PM
We precisely disrupted 3 independent meta-domains and found transcriptional defects in all 3 cases.
2 meta-domains we studied contain a meta-loop connecting a neuronal gene promoter to a distant intergenic element. Disrupting the meta-domain decreased neuronal gene transcription by 2-to-3-fold.
2 meta-domains we studied contain a meta-loop connecting a neuronal gene promoter to a distant intergenic element. Disrupting the meta-domain decreased neuronal gene transcription by 2-to-3-fold.
We recently showed that in fly neurons, specific pairs of TADs megabases apart specifically come together to form meta-domains (Mohana et al. 2023).
Within meta-domains, meta-loops connect accessible chromatin peaks present at promoters of specific neuronal genes or in non-coding intergenic DNA.
Within meta-domains, meta-loops connect accessible chromatin peaks present at promoters of specific neuronal genes or in non-coding intergenic DNA.

April 18, 2025 at 6:50 PM
We recently showed that in fly neurons, specific pairs of TADs megabases apart specifically come together to form meta-domains (Mohana et al. 2023).
Within meta-domains, meta-loops connect accessible chromatin peaks present at promoters of specific neuronal genes or in non-coding intergenic DNA.
Within meta-domains, meta-loops connect accessible chromatin peaks present at promoters of specific neuronal genes or in non-coding intergenic DNA.
and thank you to the reviewers whose feedback greatly improved the review 😎
February 13, 2025 at 12:27 PM
and thank you to the reviewers whose feedback greatly improved the review 😎
Fun to do this with talented Sanyami Zunjarrao in our lab!
Thank you @andersshansen.bsky.social and Marcelo Nollmann for including our review in the 2025 Genome Architecture & Expression edition of COGEDE 😀
Thank you @andersshansen.bsky.social and Marcelo Nollmann for including our review in the 2025 Genome Architecture & Expression edition of COGEDE 😀
February 13, 2025 at 12:26 PM
Fun to do this with talented Sanyami Zunjarrao in our lab!
Thank you @andersshansen.bsky.social and Marcelo Nollmann for including our review in the 2025 Genome Architecture & Expression edition of COGEDE 😀
Thank you @andersshansen.bsky.social and Marcelo Nollmann for including our review in the 2025 Genome Architecture & Expression edition of COGEDE 😀
yes indeed, BEAF-32 mat- zyg- mutants are not happy...
November 15, 2024 at 2:03 PM
yes indeed, BEAF-32 mat- zyg- mutants are not happy...
Thank you to the outstanding efforts of notably Anastasiia Tonelli and Pascal Cousin and another great collaboration with Aleksander Jankowski @ajank@mastodon.online 💪
November 15, 2024 at 1:59 PM
Thank you to the outstanding efforts of notably Anastasiia Tonelli and Pascal Cousin and another great collaboration with Aleksander Jankowski @ajank@mastodon.online 💪
For more info:
Open access paper: www.sciencedirect.com/science/arti... Step-by-step protocol: www.sciencedirect.com/science/arti...
Open access paper: www.sciencedirect.com/science/arti... Step-by-step protocol: www.sciencedirect.com/science/arti...

Systematic screening of enhancer-blocking insulators in Drosophila identifies their DNA sequence determinants
Long-range transcriptional activation of gene promoters by abundant enhancers in animal genomes calls for mechanisms to limit inappropriate regulation…
www.sciencedirect.com
November 15, 2024 at 1:59 PM
For more info:
Open access paper: www.sciencedirect.com/science/arti... Step-by-step protocol: www.sciencedirect.com/science/arti...
Open access paper: www.sciencedirect.com/science/arti... Step-by-step protocol: www.sciencedirect.com/science/arti...
Exciting findings using insulator-seq are:
Not all insulator-binding protein sites act as enhancer-blockers.
Not all TAD boundaries act as enhancer-blockers.
Both a CTCF motif and its surrounding sequence context are essential for enhancer-blocking.
Not all insulator-binding protein sites act as enhancer-blockers.
Not all TAD boundaries act as enhancer-blockers.
Both a CTCF motif and its surrounding sequence context are essential for enhancer-blocking.

November 15, 2024 at 1:59 PM
Exciting findings using insulator-seq are:
Not all insulator-binding protein sites act as enhancer-blockers.
Not all TAD boundaries act as enhancer-blockers.
Both a CTCF motif and its surrounding sequence context are essential for enhancer-blocking.
Not all insulator-binding protein sites act as enhancer-blockers.
Not all TAD boundaries act as enhancer-blockers.
Both a CTCF motif and its surrounding sequence context are essential for enhancer-blocking.
(2) The insulator activity of CTCF binding sites depends on an intact CTCF motif.

November 15, 2024 at 1:59 PM
(2) The insulator activity of CTCF binding sites depends on an intact CTCF motif.
Here’s some proof that insulator-seq works:
(1) It identifies biologically meaningful insulators in genomic DNA (both known and novel).
(1) It identifies biologically meaningful insulators in genomic DNA (both known and novel).

November 15, 2024 at 1:59 PM
Here’s some proof that insulator-seq works:
(1) It identifies biologically meaningful insulators in genomic DNA (both known and novel).
(1) It identifies biologically meaningful insulators in genomic DNA (both known and novel).
That’s right, insulators works on transiently transfected plasmids! This allows us to test the enhancer-blocking activities of up to 1 million DNA test fragments at a time.
November 15, 2024 at 1:59 PM
That’s right, insulators works on transiently transfected plasmids! This allows us to test the enhancer-blocking activities of up to 1 million DNA test fragments at a time.
Anastasiia Tonelli in the lab established insulator-seq - a massively parallel reporter assay for enhancer-blockers in Drosophila cells

November 15, 2024 at 1:59 PM
Anastasiia Tonelli in the lab established insulator-seq - a massively parallel reporter assay for enhancer-blockers in Drosophila cells

