Trying to master Dad mode 👨🍼
See what I'm up to here: https://github.com/mbhall88
So I rebuilt release 1 without masking, and added a release 2 database with no masking. The improvement in detection accuracy was substantial:

So I rebuilt release 1 without masking, and added a release 2 database with no masking. The improvement in detection accuracy was substantial:


LRGE uses significantly less CPU and memory than traditional approaches, making it ideal for both high-performance clusters and resource-limited environments.
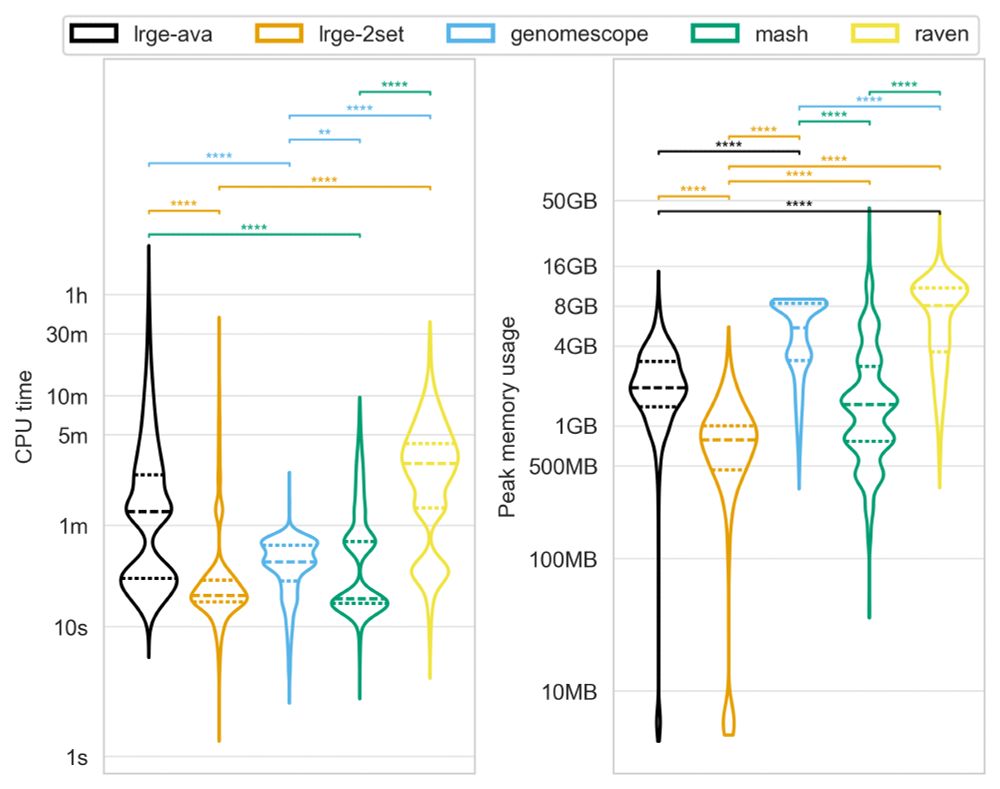
LRGE uses significantly less CPU and memory than traditional approaches, making it ideal for both high-performance clusters and resource-limited environments.
LRGE delivers estimates as reliable as assembly-based methods and better than k-mer-based approaches.
Relative error (y-axis) measures the proportional difference between the estimated and true genome size.
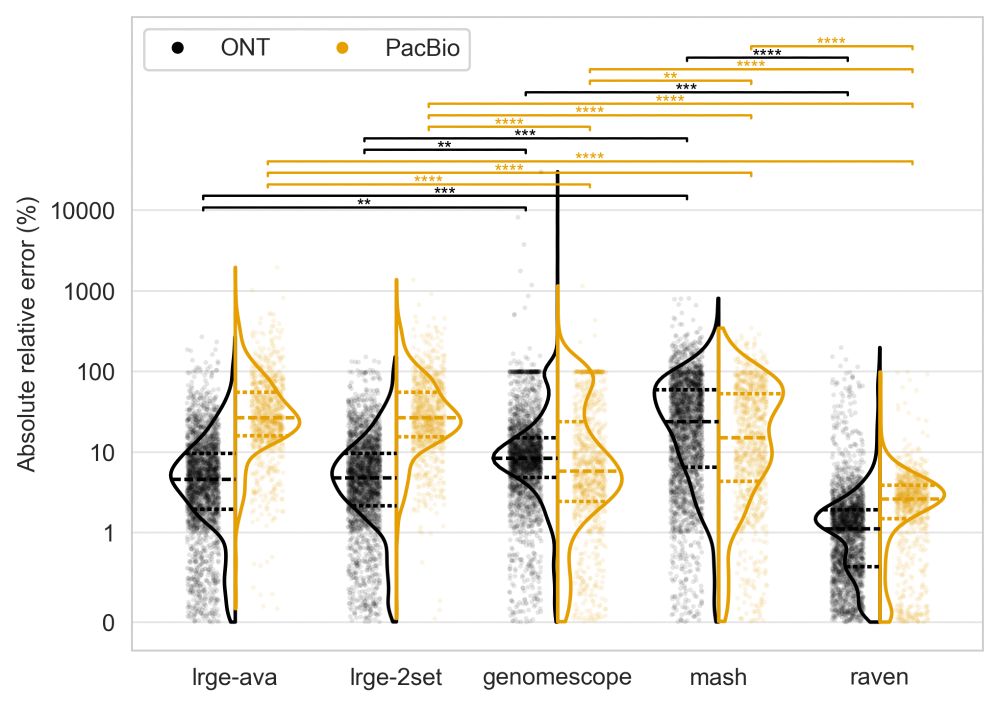
LRGE delivers estimates as reliable as assembly-based methods and better than k-mer-based approaches.
Relative error (y-axis) measures the proportional difference between the estimated and true genome size.

