
🦣 Main account: https://ecoevo.social/@marcrr
doi.org/10.1093/bios...

doi.org/10.1093/bios...

sifted.eu/articles/us-...

sifted.eu/articles/us-...






www.leslignesbougent.org/petitions/ep...

www.leslignesbougent.org/petitions/ep...


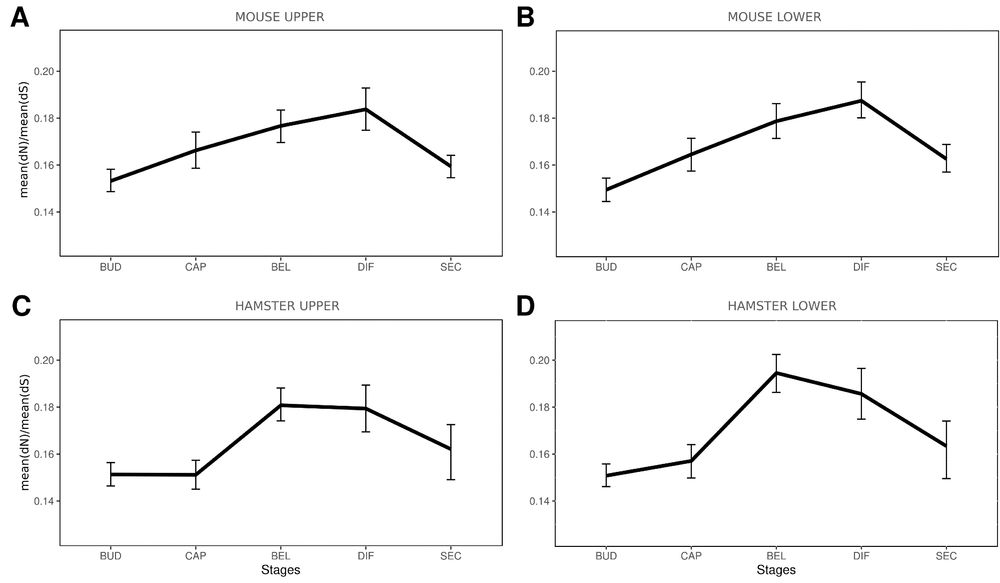
As expected from rapid molar evolution, mouse and hamster gene have rather different gene expression.
More surprising the coexpression clusters are most divergent in middle molar development.
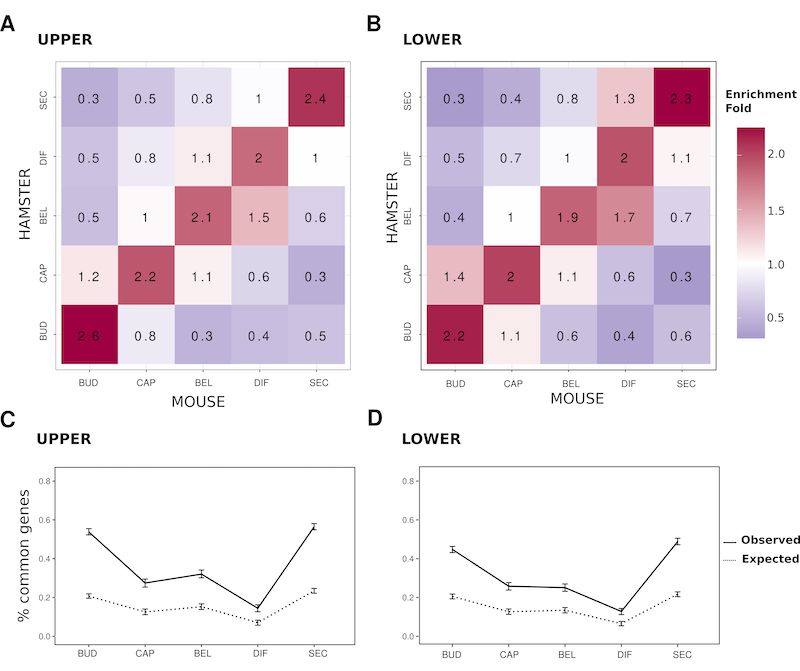
As expected from rapid molar evolution, mouse and hamster gene have rather different gene expression.
More surprising the coexpression clusters are most divergent in middle molar development.
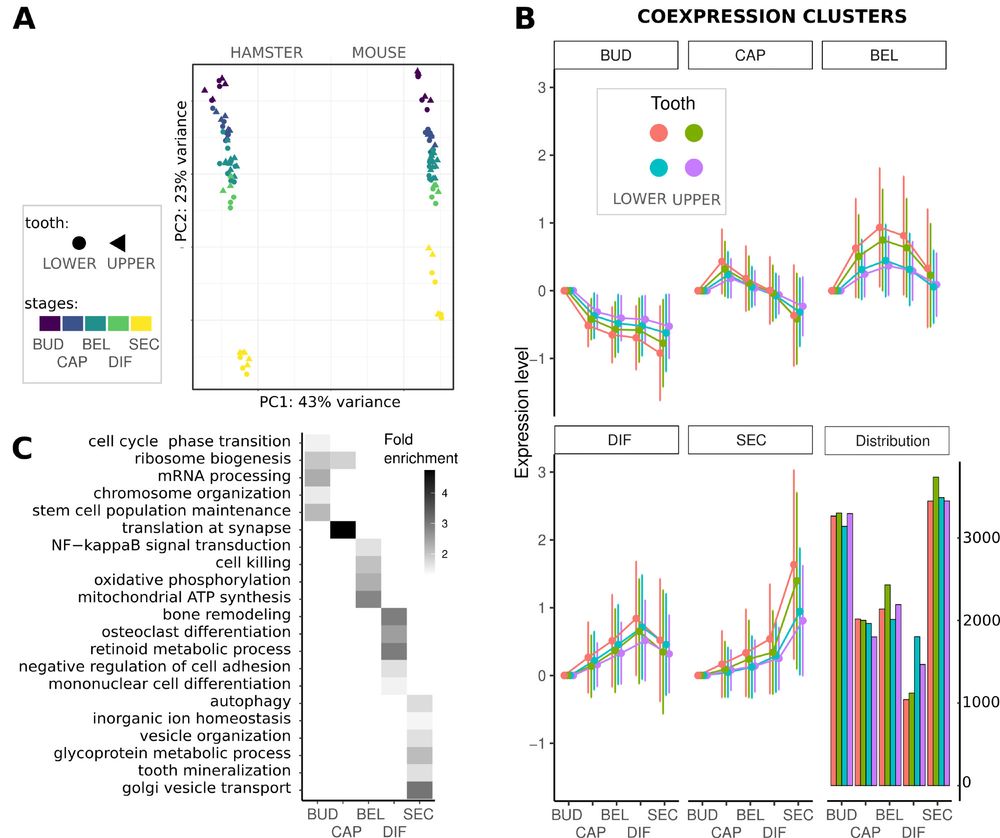
www.biorxiv.org/content/10.1...
@lbmcinlyon.bsky.social

www.biorxiv.org/content/10.1...
@lbmcinlyon.bsky.social
ecoevorxiv.org/repository/v...

ecoevorxiv.org/repository/v...

doi.org/10.1016/j.tr...

doi.org/10.1016/j.tr...

source: bouletcorp.com/rogatons/202...

source: bouletcorp.com/rogatons/202...






