Mann Lab
@mannlab.bsky.social
The Mann Lab is a pioneer in mass spectrometry-based proteomics. Posts represent personal views from lab members and Matthias Mann
If you are at HUPO 2025, catch up with our team @mannlab.bsky.social to hear about our exciting work in single-cell proteomics, immunopeptidomics, spatial proteomics, and the latest advances in mass spectrometry technology!

November 7, 2025 at 1:20 PM
If you are at HUPO 2025, catch up with our team @mannlab.bsky.social to hear about our exciting work in single-cell proteomics, immunopeptidomics, spatial proteomics, and the latest advances in mass spectrometry technology!
Awesome talk by @erictopol.bsky.social at the Bavarian Academy of Sciences and Humanities (BAdW). A positive and hopeful take on #AI in medicine. Thanks for highlighting #DVP, too 😊

October 29, 2025 at 8:58 AM
Awesome talk by @erictopol.bsky.social at the Bavarian Academy of Sciences and Humanities (BAdW). A positive and hopeful take on #AI in medicine. Thanks for highlighting #DVP, too 😊
13/
Our extensive documentation and tutorials make scPortrait easy to use and accessible.
And as part of @scverse.bsky.social it’s compatible with existing tools like scanpy, squidpy, bento-tools or Moscot 🚀🐍
mannlabs.github.io/scPortrait/i... #OpenSourceTools #Tutorial #CodeDocumentation
Our extensive documentation and tutorials make scPortrait easy to use and accessible.
And as part of @scverse.bsky.social it’s compatible with existing tools like scanpy, squidpy, bento-tools or Moscot 🚀🐍
mannlabs.github.io/scPortrait/i... #OpenSourceTools #Tutorial #CodeDocumentation
September 24, 2025 at 12:22 PM
13/
Our extensive documentation and tutorials make scPortrait easy to use and accessible.
And as part of @scverse.bsky.social it’s compatible with existing tools like scanpy, squidpy, bento-tools or Moscot 🚀🐍
mannlabs.github.io/scPortrait/i... #OpenSourceTools #Tutorial #CodeDocumentation
Our extensive documentation and tutorials make scPortrait easy to use and accessible.
And as part of @scverse.bsky.social it’s compatible with existing tools like scanpy, squidpy, bento-tools or Moscot 🚀🐍
mannlabs.github.io/scPortrait/i... #OpenSourceTools #Tutorial #CodeDocumentation
11/
We also ship a benchmark dataset of Golgi morphologies and use it to compare image featurization tools: #ConvNeXt, #SubCell, #CellProfiler
We also ship a benchmark dataset of Golgi morphologies and use it to compare image featurization tools: #ConvNeXt, #SubCell, #CellProfiler

September 24, 2025 at 12:22 PM
11/
We also ship a benchmark dataset of Golgi morphologies and use it to compare image featurization tools: #ConvNeXt, #SubCell, #CellProfiler
We also ship a benchmark dataset of Golgi morphologies and use it to compare image featurization tools: #ConvNeXt, #SubCell, #CellProfiler
10/
✨ Embedding images into transcriptome atlases ✨
We use scPortrait to embed single-cell images from a @10xgenomics.bsky.social Xenium ovarian cancer dataset into the #SCimilarity transcriptome atlas (R2 = 0.65), recovering meaningful cell types
✨ Embedding images into transcriptome atlases ✨
We use scPortrait to embed single-cell images from a @10xgenomics.bsky.social Xenium ovarian cancer dataset into the #SCimilarity transcriptome atlas (R2 = 0.65), recovering meaningful cell types

September 24, 2025 at 12:22 PM
10/
✨ Embedding images into transcriptome atlases ✨
We use scPortrait to embed single-cell images from a @10xgenomics.bsky.social Xenium ovarian cancer dataset into the #SCimilarity transcriptome atlas (R2 = 0.65), recovering meaningful cell types
✨ Embedding images into transcriptome atlases ✨
We use scPortrait to embed single-cell images from a @10xgenomics.bsky.social Xenium ovarian cancer dataset into the #SCimilarity transcriptome atlas (R2 = 0.65), recovering meaningful cell types
9/
✨ Morphology defined cell states ✨
Image embeddings generated with scPortrait resolve intra- vs extratumoral macrophages with distinct morphologies, linked to anti-inflammatory vs fibroblast-like programs
✨ Morphology defined cell states ✨
Image embeddings generated with scPortrait resolve intra- vs extratumoral macrophages with distinct morphologies, linked to anti-inflammatory vs fibroblast-like programs

September 24, 2025 at 12:22 PM
9/
✨ Morphology defined cell states ✨
Image embeddings generated with scPortrait resolve intra- vs extratumoral macrophages with distinct morphologies, linked to anti-inflammatory vs fibroblast-like programs
✨ Morphology defined cell states ✨
Image embeddings generated with scPortrait resolve intra- vs extratumoral macrophages with distinct morphologies, linked to anti-inflammatory vs fibroblast-like programs
8/
✨ Transcriptomes from images ✨
Using optimal transport + flow matching, scPortrait generates gene expression directly from CODEX images, capturing canonical marker expression like TCL1A in germinal centers in the tonsil
#CODEX #flowmatching #OT
✨ Transcriptomes from images ✨
Using optimal transport + flow matching, scPortrait generates gene expression directly from CODEX images, capturing canonical marker expression like TCL1A in germinal centers in the tonsil
#CODEX #flowmatching #OT

September 24, 2025 at 12:22 PM
8/
✨ Transcriptomes from images ✨
Using optimal transport + flow matching, scPortrait generates gene expression directly from CODEX images, capturing canonical marker expression like TCL1A in germinal centers in the tonsil
#CODEX #flowmatching #OT
✨ Transcriptomes from images ✨
Using optimal transport + flow matching, scPortrait generates gene expression directly from CODEX images, capturing canonical marker expression like TCL1A in germinal centers in the tonsil
#CODEX #flowmatching #OT
6/
The new .h5sc format provides fast random access to single-cell images for ML training.
It follows #FAIR data principles (findable, accessible, interoperable, reusable) and integrates with @scverse.bsky.social tools via AnnData.
The new .h5sc format provides fast random access to single-cell images for ML training.
It follows #FAIR data principles (findable, accessible, interoperable, reusable) and integrates with @scverse.bsky.social tools via AnnData.

September 24, 2025 at 12:22 PM
6/
The new .h5sc format provides fast random access to single-cell images for ML training.
It follows #FAIR data principles (findable, accessible, interoperable, reusable) and integrates with @scverse.bsky.social tools via AnnData.
The new .h5sc format provides fast random access to single-cell images for ML training.
It follows #FAIR data principles (findable, accessible, interoperable, reusable) and integrates with @scverse.bsky.social tools via AnnData.
5/
The scPortrait pipeline transforms raw input images step by step:
• stitch FOVs
• segment & extract cells
• output standardized .h5sc single-cell image datasets
From messy pixels → inputs ready for training 🖥️
The scPortrait pipeline transforms raw input images step by step:
• stitch FOVs
• segment & extract cells
• output standardized .h5sc single-cell image datasets
From messy pixels → inputs ready for training 🖥️

September 24, 2025 at 12:22 PM
5/
The scPortrait pipeline transforms raw input images step by step:
• stitch FOVs
• segment & extract cells
• output standardized .h5sc single-cell image datasets
From messy pixels → inputs ready for training 🖥️
The scPortrait pipeline transforms raw input images step by step:
• stitch FOVs
• segment & extract cells
• output standardized .h5sc single-cell image datasets
From messy pixels → inputs ready for training 🖥️
1/
Tomorrow’s large scale cross-modality models will further unlock biology, but they need standardized inputs. With @sophia-maedler.bsky.social & @nik-as.bsky.social, we built #scPortrait, @scverse.bsky.social package to turn microscopy images into single-cell image datasets for multimodal modeling
Tomorrow’s large scale cross-modality models will further unlock biology, but they need standardized inputs. With @sophia-maedler.bsky.social & @nik-as.bsky.social, we built #scPortrait, @scverse.bsky.social package to turn microscopy images into single-cell image datasets for multimodal modeling

September 24, 2025 at 12:22 PM
1/
Tomorrow’s large scale cross-modality models will further unlock biology, but they need standardized inputs. With @sophia-maedler.bsky.social & @nik-as.bsky.social, we built #scPortrait, @scverse.bsky.social package to turn microscopy images into single-cell image datasets for multimodal modeling
Tomorrow’s large scale cross-modality models will further unlock biology, but they need standardized inputs. With @sophia-maedler.bsky.social & @nik-as.bsky.social, we built #scPortrait, @scverse.bsky.social package to turn microscopy images into single-cell image datasets for multimodal modeling
Congratulations on the inaugural of #AITHYRA, the new Biomedical AI institute in Vienna. Thank you for the invitation to speak at the symposium and the fascinating discussions on AI × proteomics. #AIforLifeScience

September 15, 2025 at 8:10 AM
Congratulations on the inaugural of #AITHYRA, the new Biomedical AI institute in Vienna. Thank you for the invitation to speak at the symposium and the fascinating discussions on AI × proteomics. #AIforLifeScience
Withdrawal of the stem-cell niche WNR (Wnt-Noggin-R-spondin) cocktail in organoid cultures decreased proliferation and stemness, as well as improved differentiation, moving organoids toward more tissue-like states.

September 10, 2025 at 3:16 PM
Withdrawal of the stem-cell niche WNR (Wnt-Noggin-R-spondin) cocktail in organoid cultures decreased proliferation and stemness, as well as improved differentiation, moving organoids toward more tissue-like states.
scDVP’s high-resolution enabled us to observe fine-grained protein abundance changes along the colon crypt axis. For instance, the abundance gradient of CA1 was only visible in organoid transplants and in vivo tissue.

September 10, 2025 at 3:16 PM
scDVP’s high-resolution enabled us to observe fine-grained protein abundance changes along the colon crypt axis. For instance, the abundance gradient of CA1 was only visible in organoid transplants and in vivo tissue.
We further employed scDVP to explore differences in individual intestinal epithelial cells. This dataset comprised about 2,700 proteins across cell types, and confirmed clearer stemness-to-differentiation programs after organoid xenotransplantation.

September 10, 2025 at 3:16 PM
We further employed scDVP to explore differences in individual intestinal epithelial cells. This dataset comprised about 2,700 proteins across cell types, and confirmed clearer stemness-to-differentiation programs after organoid xenotransplantation.
Led by @freddyomics.bsky.social and @hausmannannika.bsky.social, we created a spatial human colon resource consisting of almost 8,000 protein groups across distinct cell types! This reference enabled us to assess cell populations of human colon organoids and establish a colon stemness signature.

September 10, 2025 at 3:16 PM
Led by @freddyomics.bsky.social and @hausmannannika.bsky.social, we created a spatial human colon resource consisting of almost 8,000 protein groups across distinct cell types! This reference enabled us to assess cell populations of human colon organoids and establish a colon stemness signature.
Out in @cp-cellsystems.bsky.social: Deep Visual Proteomics shows xenotransplantation drives colon #organoids toward in-vivo-like proteomes supporting their use in regenerative medicine; culture tweaks can mimic this shift. With @kimbakjensen.bsky.social. Lead author Frederik Post explains below:

September 10, 2025 at 3:16 PM
Out in @cp-cellsystems.bsky.social: Deep Visual Proteomics shows xenotransplantation drives colon #organoids toward in-vivo-like proteomes supporting their use in regenerative medicine; culture tweaks can mimic this shift. With @kimbakjensen.bsky.social. Lead author Frederik Post explains below:
If you are at the ASBMB Proteomics conference next week, do attend the session talks and lunch seminars
@abseabio.bsky.social, @evosep.bsky.social to know about the latest research developments in @mannlab.bsky.social
@abseabio.bsky.social, @evosep.bsky.social to know about the latest research developments in @mannlab.bsky.social

August 13, 2025 at 9:21 AM
If you are at the ASBMB Proteomics conference next week, do attend the session talks and lunch seminars
@abseabio.bsky.social, @evosep.bsky.social to know about the latest research developments in @mannlab.bsky.social
@abseabio.bsky.social, @evosep.bsky.social to know about the latest research developments in @mannlab.bsky.social
#6: Our integrative approach led to validation of 16 therapeutic targets. Combining Milciclib (CDK inhibitor) with Mirvetuximab (targets FOLR1) significantly reduced tumor burden in vivo—a promising new strategy for treating chemo-resistant LGSC.
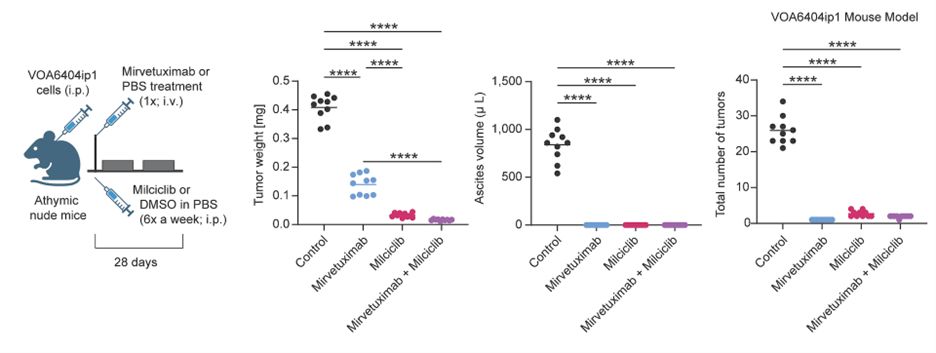
June 26, 2025 at 3:20 PM
#6: Our integrative approach led to validation of 16 therapeutic targets. Combining Milciclib (CDK inhibitor) with Mirvetuximab (targets FOLR1) significantly reduced tumor burden in vivo—a promising new strategy for treating chemo-resistant LGSC.
#5: For clinical interpretation, we performed a multi-modal data integration – ranging from clinical pathology to the molecular fingerprint of each sample.

June 26, 2025 at 3:20 PM
#5: For clinical interpretation, we performed a multi-modal data integration – ranging from clinical pathology to the molecular fingerprint of each sample.
#4: Spatial mapping of tumor–stroma interactions revealed how epithelial tumor cells engage their environment—highlighting key roles for integrins and fibronectin in facilitating invasion and matrix remodeling.

June 26, 2025 at 3:20 PM
#4: Spatial mapping of tumor–stroma interactions revealed how epithelial tumor cells engage their environment—highlighting key roles for integrins and fibronectin in facilitating invasion and matrix remodeling.
#3: Our proteomic profiling of epithelial cells revealed several key mechanisms, including the exclusive expression of the brain-specific splicing regulator NOVA in invasive cancer. A molecular ‘switch’ to invasive tumor growth?

June 26, 2025 at 3:20 PM
#3: Our proteomic profiling of epithelial cells revealed several key mechanisms, including the exclusive expression of the brain-specific splicing regulator NOVA in invasive cancer. A molecular ‘switch’ to invasive tumor growth?
#2: Using cell-type resolved #DeepVisualProteomics, we uncovered a gradual transformation from SBT to LGSC, via micropapillary tumors as a transition state—profiling up to 5,400 proteins from just ~200 cells per sample!

June 26, 2025 at 3:20 PM
#2: Using cell-type resolved #DeepVisualProteomics, we uncovered a gradual transformation from SBT to LGSC, via micropapillary tumors as a transition state—profiling up to 5,400 proteins from just ~200 cells per sample!
#1: Low-grade serous ovarian cancer (LGSC) is a chemo-resistant and metastasizing disease that affects younger women. We studied its origin by analyzing the progression from serous borderline tumors (SBT) across epithelial and stromal cells.

June 26, 2025 at 3:20 PM
#1: Low-grade serous ovarian cancer (LGSC) is a chemo-resistant and metastasizing disease that affects younger women. We studied its origin by analyzing the progression from serous borderline tumors (SBT) across epithelial and stromal cells.
Don't forget to check out Marvin Thielert's poster 773 on single cell proteomics at #ASMS2025! #SingleCellProteomics

June 4, 2025 at 2:02 PM
Don't forget to check out Marvin Thielert's poster 773 on single cell proteomics at #ASMS2025! #SingleCellProteomics
Nano-scale, mega-impact: nanoPhos delivers 100× phosphoproteomics sensitivity, 70k sites from 1µg, 8k from 10ng! Powered by SPEC, it enables phosphoDVP: deep spatial phosphoproteomics from 1000 neurons, hitting 17k sites, a game changer for spatial omics.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

June 2, 2025 at 9:49 AM
Nano-scale, mega-impact: nanoPhos delivers 100× phosphoproteomics sensitivity, 70k sites from 1µg, 8k from 10ng! Powered by SPEC, it enables phosphoDVP: deep spatial phosphoproteomics from 1000 neurons, hitting 17k sites, a game changer for spatial omics.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

