Malte Kuehl
@maltekuehl.com
Computational medicine @ AU
Founder @ KH Biotechnology
Creator of spatiomic, pytximport and https://biocontext.ai
Interested in spatial omics, cancer & ageing
Lab: https://github.com/complextissue/
Personal: https://maltekuehl.com
Opinions my own.
Founder @ KH Biotechnology
Creator of spatiomic, pytximport and https://biocontext.ai
Interested in spatial omics, cancer & ageing
Lab: https://github.com/complextissue/
Personal: https://maltekuehl.com
Opinions my own.
You can also easily add it to existing chatbots like Claude Desktop, VS Code and others. Find out how in our README: github.com/biocontext-a...

July 29, 2025 at 12:41 PM
You can also easily add it to existing chatbots like Claude Desktop, VS Code and others. Find out how in our README: github.com/biocontext-a...
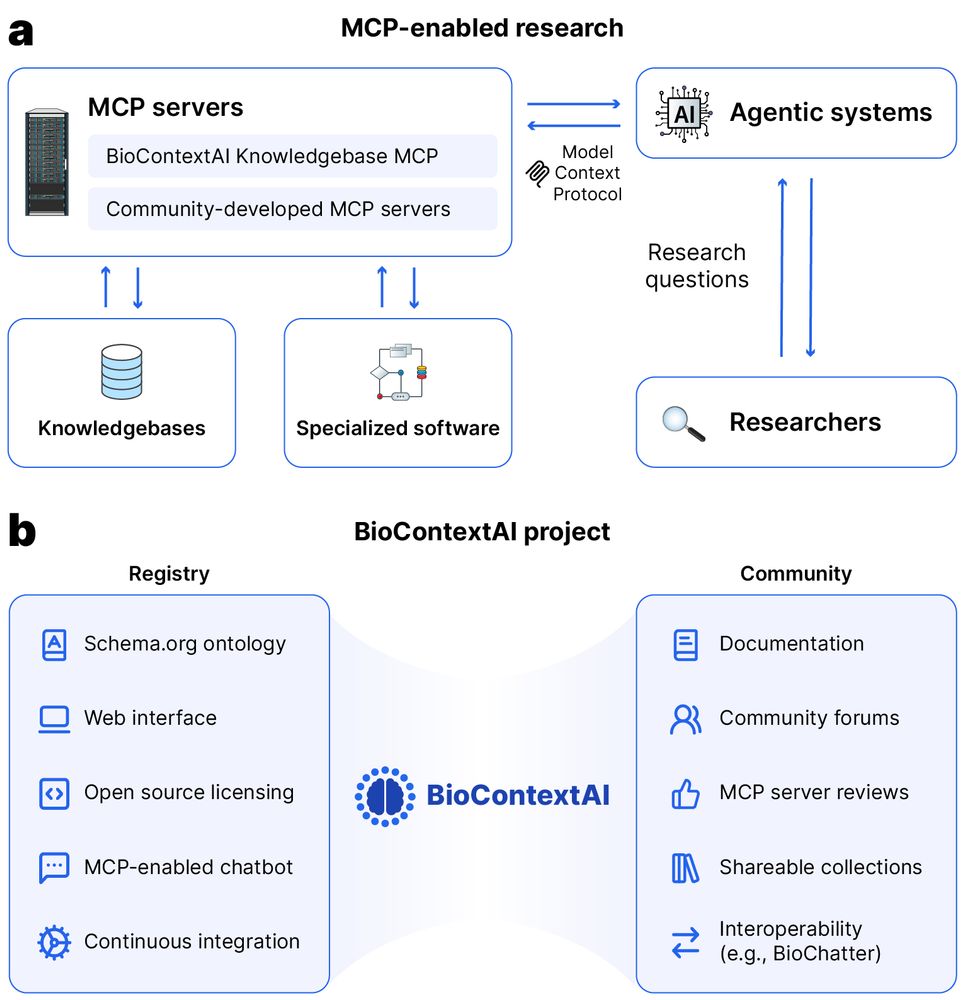
If you would like to explore how MCP servers might be helpful for your work, we provide a reference server called "Knowledgebase MCP" that integrates UniProt, STRING, PRIDE, InterPro, ClinicalTrials, EuropePMC and many more sources. You can start chatting at: biocontext.ai/chat

July 29, 2025 at 12:41 PM
If you would like to explore how MCP servers might be helpful for your work, we provide a reference server called "Knowledgebase MCP" that integrates UniProt, STRING, PRIDE, InterPro, ClinicalTrials, EuropePMC and many more sources. You can start chatting at: biocontext.ai/chat
Preprint alert 🚨 Do you use chatbots in your work or even build MCP servers and agentic systems yourself?
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
July 29, 2025 at 12:41 PM
Preprint alert 🚨 Do you use chatbots in your work or even build MCP servers and agentic systems yourself?
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
And even before histologically or clinically detectable disease, PathoPlex was able to detect several changes at the molecular level in young adults with early-onset type 2 diabetes and profile the effect of SGLT2i administration on renal metabolism and signalling.

July 18, 2025 at 1:31 PM
And even before histologically or clinically detectable disease, PathoPlex was able to detect several changes at the molecular level in young adults with early-onset type 2 diabetes and profile the effect of SGLT2i administration on renal metabolism and signalling.
In a cohort of patients with advanced diabetic kidney disease, our antibody panel of 61 markers led to the identification of tubular calcium signalling as a potential target for pharmacological intervention 💊

July 18, 2025 at 1:31 PM
In a cohort of patients with advanced diabetic kidney disease, our antibody panel of 61 markers led to the identification of tubular calcium signalling as a potential target for pharmacological intervention 💊
In a multimodal cross species validation experiment guided by PathoPlex, we were able to characterize pJUN mediated signalling in PECs as a key feature of disease.

July 18, 2025 at 1:31 PM
In a multimodal cross species validation experiment guided by PathoPlex, we were able to characterize pJUN mediated signalling in PECs as a key feature of disease.
A key use case unlocked by spatiomic is pixel-level clustering of protein co-expression patterns. This not only looks beautiful, but combined with confocal microscopy also enables truly subcellular image analysis 🖥️

July 18, 2025 at 1:31 PM
A key use case unlocked by spatiomic is pixel-level clustering of protein co-expression patterns. This not only looks beautiful, but combined with confocal microscopy also enables truly subcellular image analysis 🖥️
On the image analysis side, we developed a new GPU-accelerated Python package termed "spatiomic". It is available through PyPi with documentation and a full example notebook available at: spatiomic.org
Code is available at: github.com/complextissu...
Code is available at: github.com/complextissu...

July 18, 2025 at 1:31 PM
On the image analysis side, we developed a new GPU-accelerated Python package termed "spatiomic". It is available through PyPi with documentation and a full example notebook available at: spatiomic.org
Code is available at: github.com/complextissu...
Code is available at: github.com/complextissu...
Our imaging protocol builds on 4i (Gut et al., 2018) but scales it to > 100 proteins in pathology specimens. We optimise tissue stability through the use of APTES as a coating agent and achieve high throughput through 3D-printed tissue chambers that can fit 40+ samples and prevent pipetting errors.

July 18, 2025 at 1:31 PM
Our imaging protocol builds on 4i (Gut et al., 2018) but scales it to > 100 proteins in pathology specimens. We optimise tissue stability through the use of APTES as a coating agent and achieve high throughput through 3D-printed tissue chambers that can fit 40+ samples and prevent pipetting errors.
PathoPlex represents a flexible framework that leverages off-the-shelf antibodies and covers both multiplex imaging as well as image analysis at a subcellular resolution.
Let's go through the parts 🚀
Let's go through the parts 🚀

July 18, 2025 at 1:31 PM
PathoPlex represents a flexible framework that leverages off-the-shelf antibodies and covers both multiplex imaging as well as image analysis at a subcellular resolution.
Let's go through the parts 🚀
Let's go through the parts 🚀
Of course, you can also use pytximport from within Python with an easy-to-learn API. (8/12)
![Python code to use pytximport:
from pytximport import tximport
from pytximport.utils import create_transcript_to_gene_map
tx2gene = create_transcript_to_gene_map(
species="human".
target_field="external_gene_name",
)
txi = tximport(
[" ./quant.sf"],
data_type="salmon",
transcript_gene_map=tx2gene,
counts_from_abundance="length_scaled_tpm",
)](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:rznwyvucajdzngmwtpgkzyjy/bafkreibnnns6mkmjatl6mn2pki2oxaimuejrqda6gy45pkvy6ite2wra4u@jpeg)
November 29, 2024 at 5:24 PM
Of course, you can also use pytximport from within Python with an easy-to-learn API. (8/12)
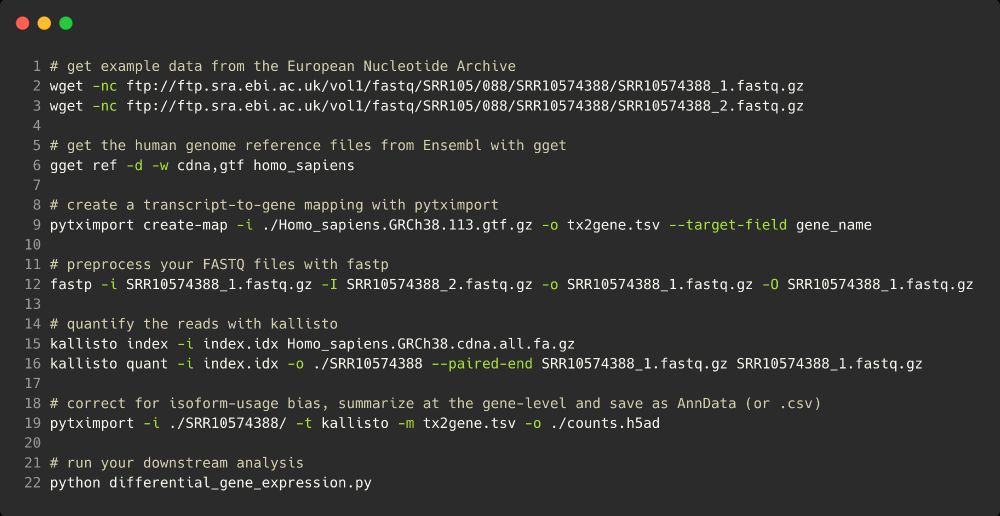
With pytximport and tools like fastp by Shifu Chen, kallisto by the @lpachter.bsky.social lab and gget by @lauraluebbert.com et al, running a bulk RNA-sequencing analysis is now a 10 line bash script. (7/12)
November 29, 2024 at 5:24 PM
With pytximport and tools like fastp by Shifu Chen, kallisto by the @lpachter.bsky.social lab and gget by @lauraluebbert.com et al, running a bulk RNA-sequencing analysis is now a 10 line bash script. (7/12)
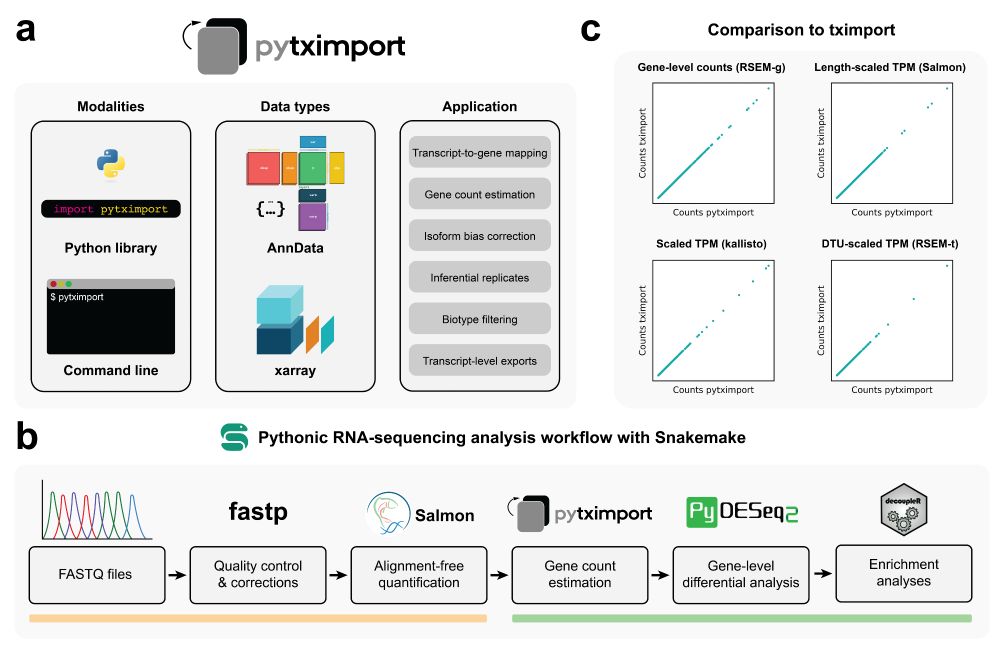
Coding in Python and looking to perform bulk RNA sequencing analysis? With pytximport recently published in Bioinformatics and version 0.11.0 out today with many improvements, it’s time for my first Bluetorial!
A thread 🧵 (1/12)
A thread 🧵 (1/12)
November 29, 2024 at 5:24 PM
Coding in Python and looking to perform bulk RNA sequencing analysis? With pytximport recently published in Bioinformatics and version 0.11.0 out today with many improvements, it’s time for my first Bluetorial!
A thread 🧵 (1/12)
A thread 🧵 (1/12)

