🧵 16/n
🧵 16/n
🧵 15/n
🧵 15/n
That’s it! Thanks for sticking with me through this thread!
🧵 14/n
That’s it! Thanks for sticking with me through this thread!
🧵 14/n
🧵 13/n

🧵 13/n
🧵 12/n

🧵 12/n
We then used high performing phenotypes to annotate our corpus of >1.3M genomes with 32 diverse phenotypic traits.
🧵 11/n
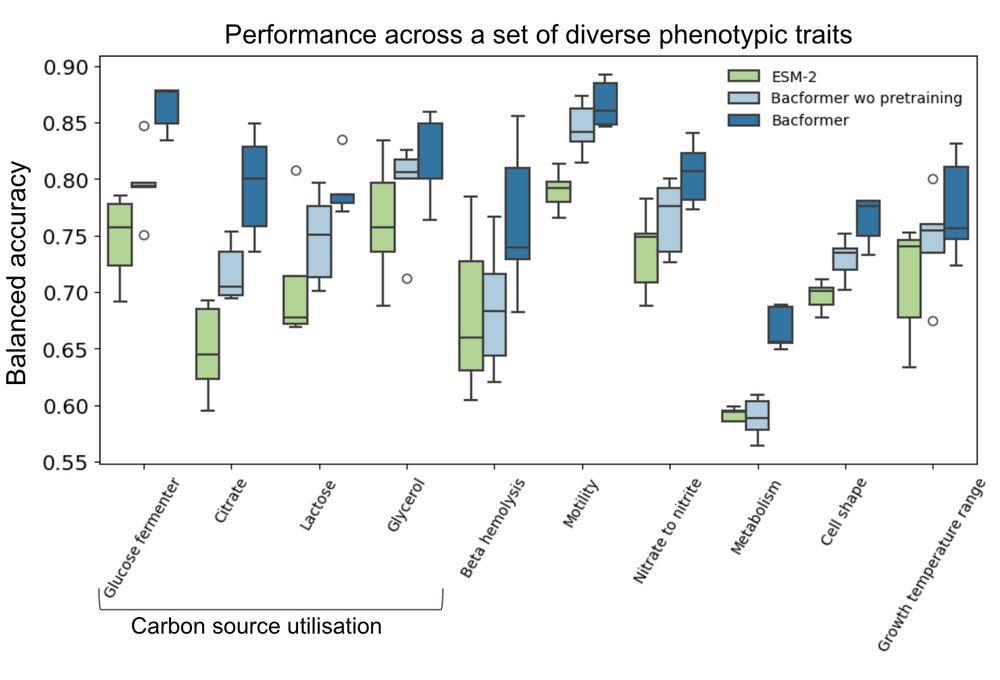
We then used high performing phenotypes to annotate our corpus of >1.3M genomes with 32 diverse phenotypic traits.
🧵 11/n
To us, this make sense as gene's essentiality and function is often tied with its genomic neighborhoud in bacteria.
🧵 10/n

To us, this make sense as gene's essentiality and function is often tied with its genomic neighborhoud in bacteria.
🧵 10/n
To do it, we fine-tuned the model on STRING DB and used it to predict the interactome of P. aeruginosa, with the top scoring pairs showing high-confidence interfaces based on AF3.
🧵 9/n

To do it, we fine-tuned the model on STRING DB and used it to predict the interactome of P. aeruginosa, with the top scoring pairs showing high-confidence interfaces based on AF3.
🧵 9/n
🧵 8/n

🧵 8/n
We examined whether Bacformer can uncover the evolutionary relationships by examining if the genome embeddings from Bacformer can be used for clustering without any "species" token.
🧵 7/n

We examined whether Bacformer can uncover the evolutionary relationships by examining if the genome embeddings from Bacformer can be used for clustering without any "species" token.
🧵 7/n
🧵 6/n

🧵 6/n
🧵 5/n

🧵 5/n
🧵 4/n

🧵 4/n
🧵 3/n
🧵 3/n
Blog ✍️: macwiatrak.github.io/posts/2025/i...
Pretrained weights 🤖: huggingface.co/macwiatrak
🧵 2/n

Blog ✍️: macwiatrak.github.io/posts/2025/i...
Pretrained weights 🤖: huggingface.co/macwiatrak
🧵 2/n

