🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
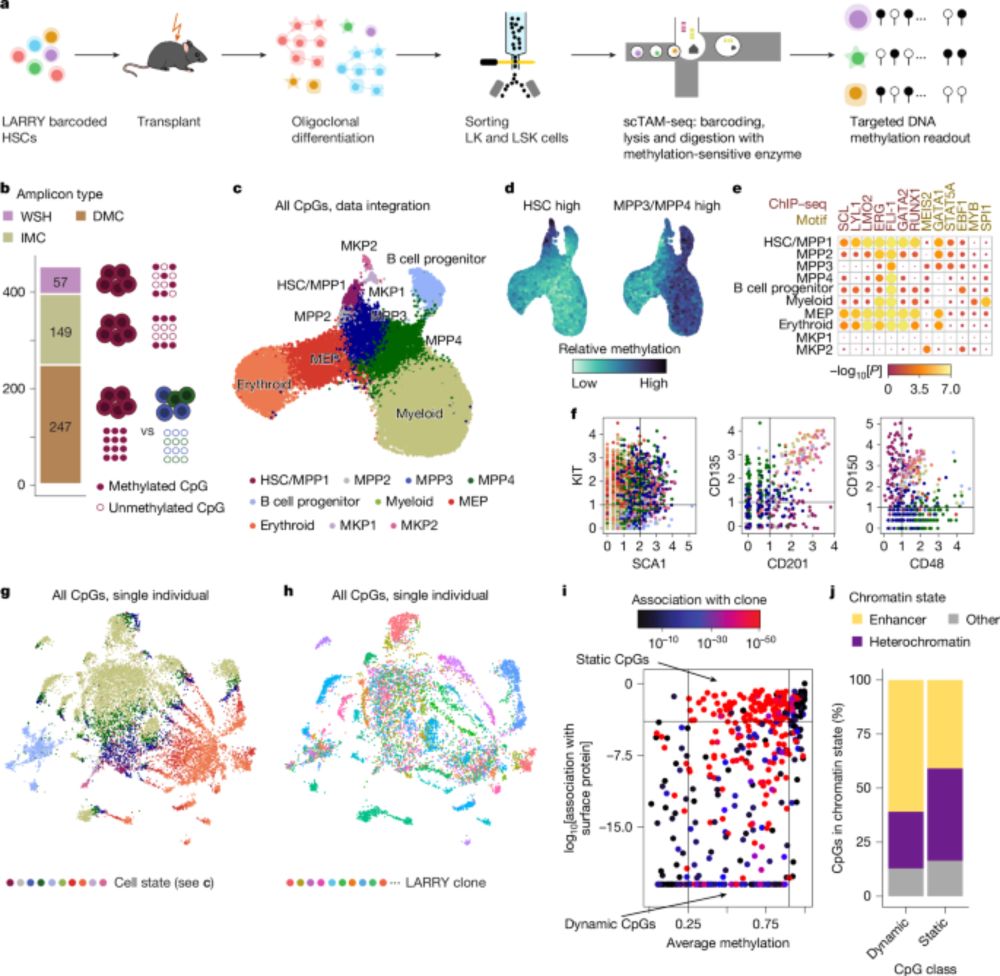
I loved analyzing the human data - we discovered that after age 50, blood production shifts to an oligoclonal pattern, and these expansions aren’t driven by CH mutations!
Dive into @larsplus.bsky.social thread for the full story & visuals.
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵

I loved analyzing the human data - we discovered that after age 50, blood production shifts to an oligoclonal pattern, and these expansions aren’t driven by CH mutations!
Dive into @larsplus.bsky.social thread for the full story & visuals.

