
Lorenz Lamm
@lorenzlamm.bsky.social
PhD Student at Helmholtz AI | MemBrain analysis for Cryo-ET
#teamTomo
#teamTomo
🔑 Usability
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
April 25, 2025 at 7:28 AM
🔑 Usability
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
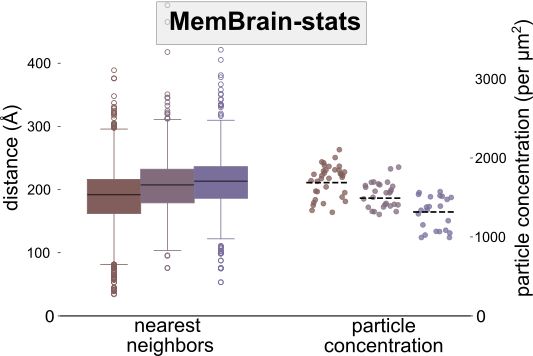
⚖️ MemBrain-stats
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
April 25, 2025 at 7:28 AM
⚖️ MemBrain-stats
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
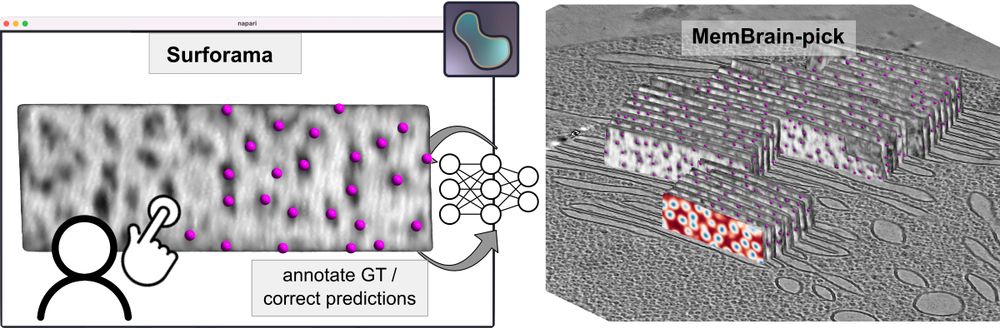
⛏️MemBrain-pick
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
April 25, 2025 at 7:28 AM
⛏️MemBrain-pick
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
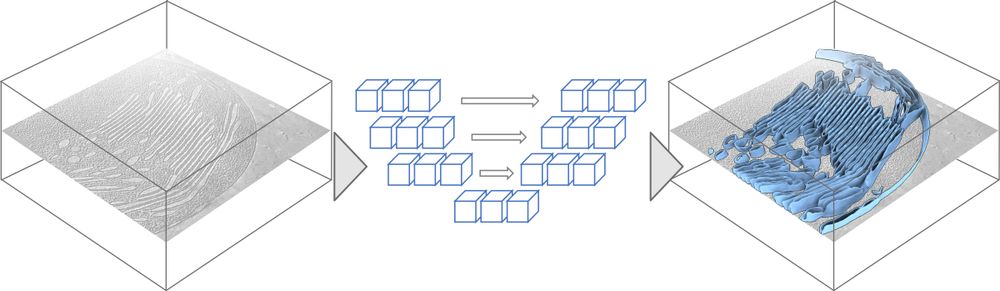
🎨 MemBrain-seg
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)
April 25, 2025 at 7:28 AM
🎨 MemBrain-seg
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)
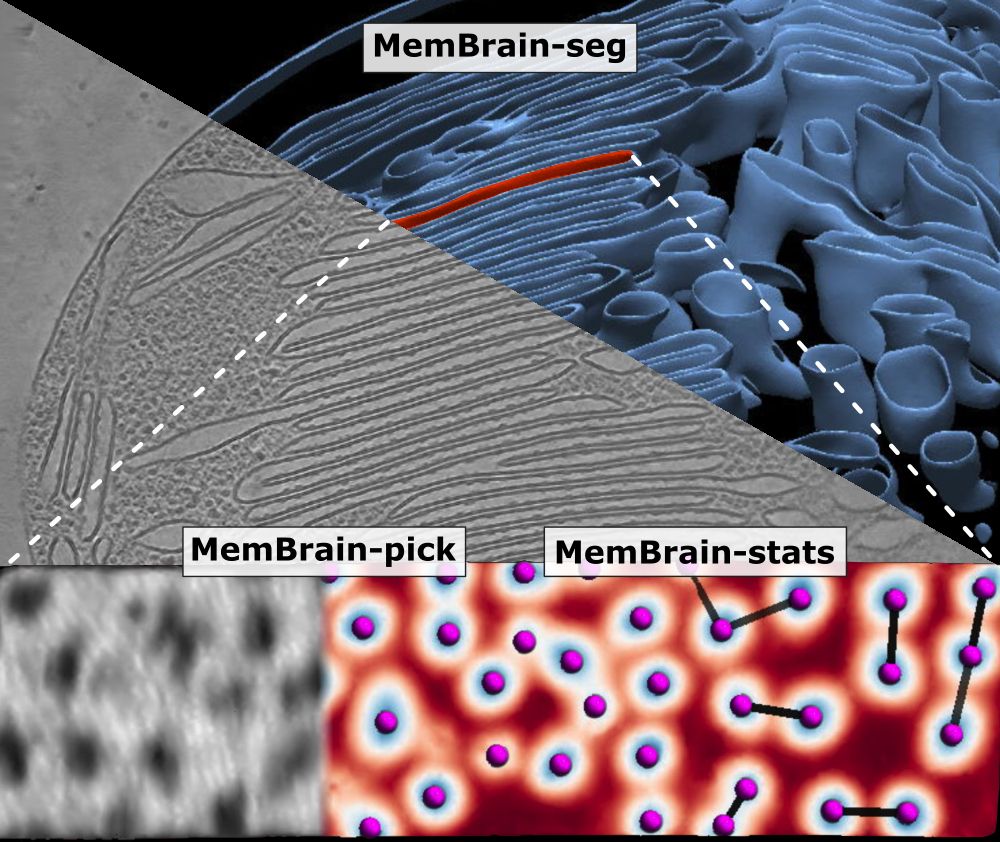
🦠🧠 MemBrain update! 🧠🦠
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
April 25, 2025 at 7:28 AM
🦠🧠 MemBrain update! 🧠🦠
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo

