S. Lorena Ament
@loreament.bsky.social
Evolutionary biologist interested in genomics, speciation, reproductive strategies, fungi, and biodiversity. And cats! She/her
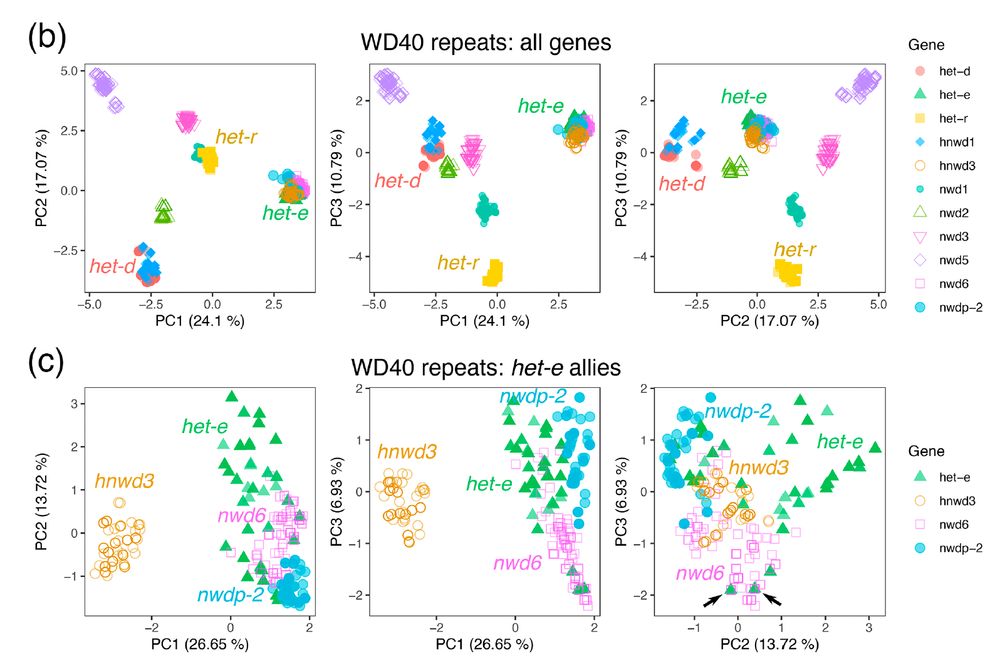
For the repeat domain, we extracted >1100 individual repeats and performed a PCA of their nucleotide sequences. Indeed, het-d and het-e are not that similar, but we discovered that het-e seems to be exchanging repeats with another gene! 8/n
July 2, 2025 at 6:10 PM
For the repeat domain, we extracted >1100 individual repeats and performed a PCA of their nucleotide sequences. Indeed, het-d and het-e are not that similar, but we discovered that het-e seems to be exchanging repeats with another gene! 8/n
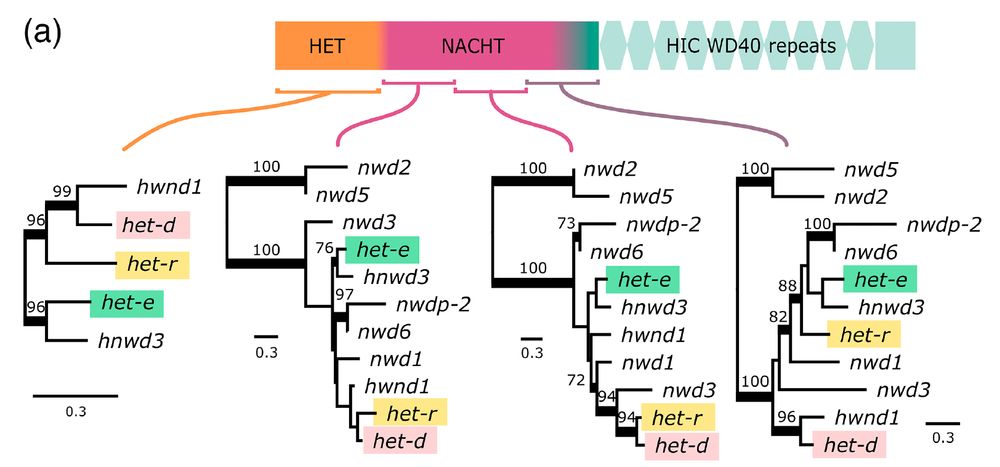
Are het-d and het-e capable of recognizing het-c because of shared ancestry? No! It turns out they are not that closely related, judging by phylogenies along the gene for a bunch of NLRs in Podospora. 7/n
July 2, 2025 at 6:10 PM
Are het-d and het-e capable of recognizing het-c because of shared ancestry? No! It turns out they are not that closely related, judging by phylogenies along the gene for a bunch of NLRs in Podospora. 7/n
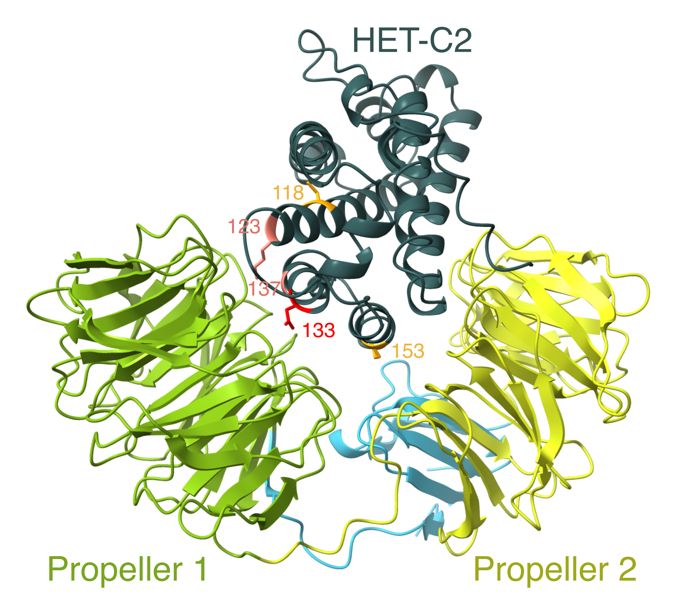
We sequenced a panel of strains with known alleles, which can now be used to assign a phenotype to wild-type strains. With this information in hand, we determined that reactive alleles fold into two beta-propellers that likely embrace their cognate ligand HET-C like a clam. 6/n
#AlphaFold
#AlphaFold
July 2, 2025 at 6:10 PM
We sequenced a panel of strains with known alleles, which can now be used to assign a phenotype to wild-type strains. With this information in hand, we determined that reactive alleles fold into two beta-propellers that likely embrace their cognate ligand HET-C like a clam. 6/n
#AlphaFold
#AlphaFold
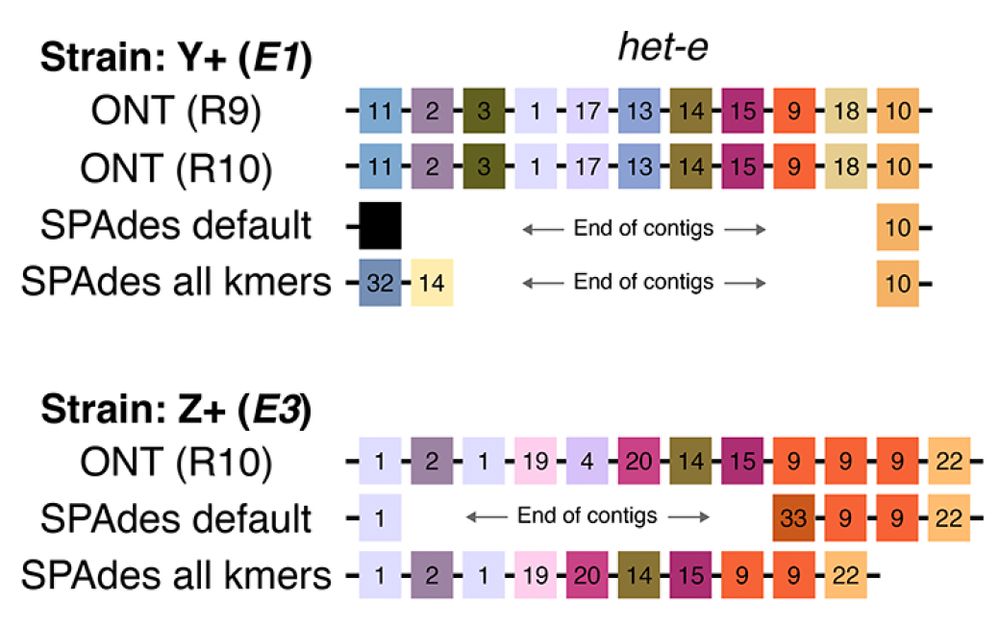
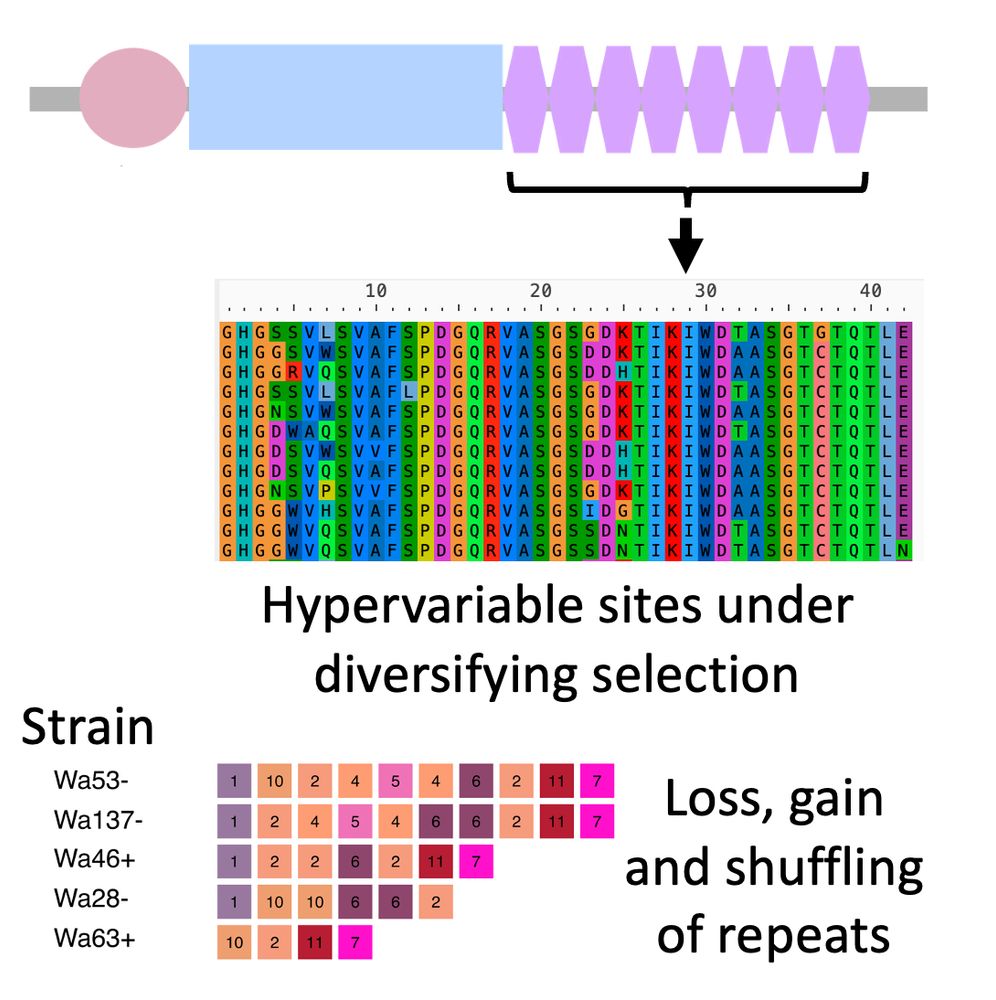
het-d/e have "high internal conservation" (HIC): their C-term domain is made out of nearly identical WD40 repeats. Only a few sites are different and evolve under diversifying selection. Concerted evolution (or something) homogenizes the repeats and leads to loss, gain, and shuffling of repeats. 4/n
July 2, 2025 at 6:10 PM
het-d/e have "high internal conservation" (HIC): their C-term domain is made out of nearly identical WD40 repeats. Only a few sites are different and evolve under diversifying selection. Concerted evolution (or something) homogenizes the repeats and leads to loss, gain, and shuffling of repeats. 4/n
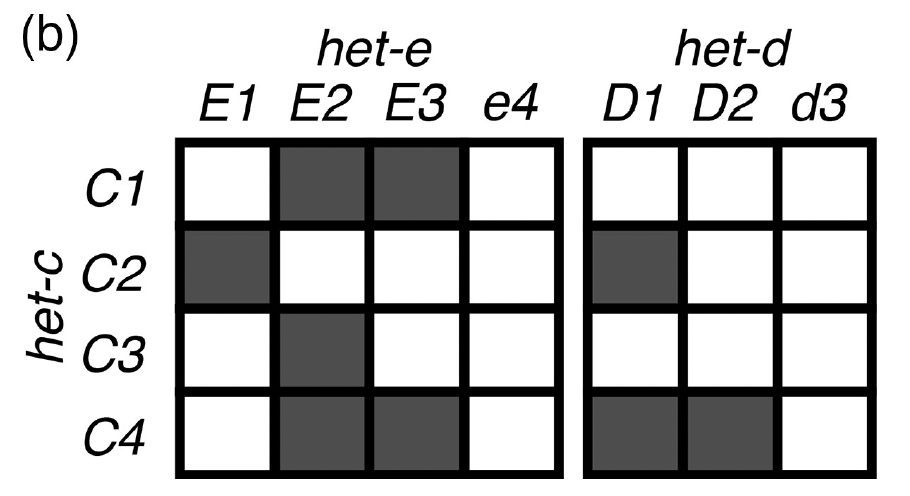
In the model fungus #Podospora anserina, the het-d
and het-e NLRs are highly polymorphic genes, whose products recognize different alleles of another gene, het-c. If two confronting strains have incompatible het-e/d and het-c alleles, a rejection reaction occurs. 3/n
and het-e NLRs are highly polymorphic genes, whose products recognize different alleles of another gene, het-c. If two confronting strains have incompatible het-e/d and het-c alleles, a rejection reaction occurs. 3/n
July 2, 2025 at 6:10 PM
In the model fungus #Podospora anserina, the het-d
and het-e NLRs are highly polymorphic genes, whose products recognize different alleles of another gene, het-c. If two confronting strains have incompatible het-e/d and het-c alleles, a rejection reaction occurs. 3/n
and het-e NLRs are highly polymorphic genes, whose products recognize different alleles of another gene, het-c. If two confronting strains have incompatible het-e/d and het-c alleles, a rejection reaction occurs. 3/n
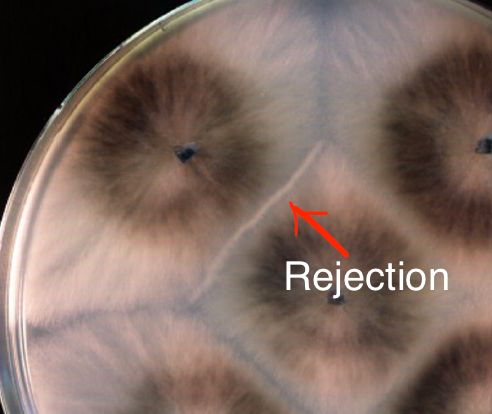
NOD-like receptors (NLRs) are intracellular proteins that play key roles in the innate immune system of plants and animals. But in fungi we know the function of only a few cases: they all work in heterokaryon incompatibility (rejection between different strains during vegetative fusion). 2/n
July 2, 2025 at 6:10 PM
NOD-like receptors (NLRs) are intracellular proteins that play key roles in the innate immune system of plants and animals. But in fungi we know the function of only a few cases: they all work in heterokaryon incompatibility (rejection between different strains during vegetative fusion). 2/n