AMR | Campylobacter | Pathogen genomics
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
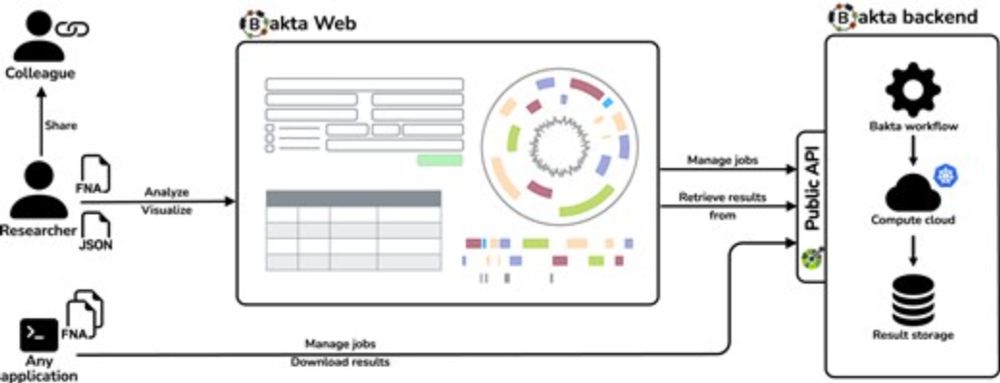
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
After a year, it was time for a Bakta database update - and it's a huge one:
- IPS: 330.9M
- PSC: 135.3M
- PSCC: 37M
doi.org/10.5281/zeno...
👇 1/6
After a year, it was time for a Bakta database update - and it's a huge one:
- IPS: 330.9M
- PSC: 135.3M
- PSCC: 37M
doi.org/10.5281/zeno...
👇 1/6
We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)

We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)
www.werkenbijumcutrecht.nl/vacatures/we...

www.werkenbijumcutrecht.nl/vacatures/we...
forms.gle/YJP6WQjsk8KK...

forms.gle/YJP6WQjsk8KK...

