
Dr Liam Brierley
@liambrierley.bsky.social
Virologist, statistician, and science presenter.
Runs @vibelab.co.uk
Research Fellow at @cvrinfo.bsky.social
Ambassador for @royalstatsoc.bsky.social
Five parts emerging virus epi, two parts R/compsci, ten parts caffeine.
he/him 🏳️🌈♾
Runs @vibelab.co.uk
Research Fellow at @cvrinfo.bsky.social
Ambassador for @royalstatsoc.bsky.social
Five parts emerging virus epi, two parts R/compsci, ten parts caffeine.
he/him 🏳️🌈♾
@besaldernetwork.bsky.social
@qedmathsnetwork.bsky.social
@lgbtmath.org
@outtoinnovate.bsky.social
@lgbtqstem.bsky.social
Please let me know any other networks we should share this with!
@qedmathsnetwork.bsky.social
@lgbtmath.org
@outtoinnovate.bsky.social
@lgbtqstem.bsky.social
Please let me know any other networks we should share this with!
November 11, 2025 at 4:56 PM
@besaldernetwork.bsky.social
@qedmathsnetwork.bsky.social
@lgbtmath.org
@outtoinnovate.bsky.social
@lgbtqstem.bsky.social
Please let me know any other networks we should share this with!
@qedmathsnetwork.bsky.social
@lgbtmath.org
@outtoinnovate.bsky.social
@lgbtqstem.bsky.social
Please let me know any other networks we should share this with!
You can when smartass titles become a tickbox in the CRediT author taxonomy 😁
September 19, 2025 at 10:37 AM
You can when smartass titles become a tickbox in the CRediT author taxonomy 😁
This is the 2nd preprint to come from collaboration between @thepandemicinst.bsky.social @livuniresearch.bsky.social & CSL Seqirus, supported by @baylism.bsky.social & Joaquin Mould.
We hope this work can help risk assess new bird flu strains and flag key mutations in the wild!
#preprint #avianflu
We hope this work can help risk assess new bird flu strains and flag key mutations in the wild!
#preprint #avianflu
September 18, 2025 at 1:39 PM
This is the 2nd preprint to come from collaboration between @thepandemicinst.bsky.social @livuniresearch.bsky.social & CSL Seqirus, supported by @baylism.bsky.social & Joaquin Mould.
We hope this work can help risk assess new bird flu strains and flag key mutations in the wild!
#preprint #avianflu
We hope this work can help risk assess new bird flu strains and flag key mutations in the wild!
#preprint #avianflu
This stack is able to correctly predict zoonotic potential of sequences in entirely unseen subtypes with AUC=0.95 and F1=0.90, a level of generalisability that is not often seen for machine learning host predictors.
Interestingly, it flags some duck H4 viruses from Americas as having distinct risk.
Interestingly, it flags some duck H4 viruses from Americas as having distinct risk.

September 18, 2025 at 1:39 PM
This stack is able to correctly predict zoonotic potential of sequences in entirely unseen subtypes with AUC=0.95 and F1=0.90, a level of generalisability that is not often seen for machine learning host predictors.
Interestingly, it flags some duck H4 viruses from Americas as having distinct risk.
Interestingly, it flags some duck H4 viruses from Americas as having distinct risk.
Training on 12 feature sets over each of 8 segments, we find protein properties are usually best at estimating zoonotic potential from a single segment.
But what about whole genomes? We can combine the best models in a single trained meta-learner (or "stack"), that draws on info from all of them!
But what about whole genomes? We can combine the best models in a single trained meta-learner (or "stack"), that draws on info from all of them!

September 18, 2025 at 1:39 PM
Training on 12 feature sets over each of 8 segments, we find protein properties are usually best at estimating zoonotic potential from a single segment.
But what about whole genomes? We can combine the best models in a single trained meta-learner (or "stack"), that draws on info from all of them!
But what about whole genomes? We can combine the best models in a single trained meta-learner (or "stack"), that draws on info from all of them!
We extracted ~19000 influenza sequences from birds and ~600 zoonotic sequences from humans (only non-seasonal subtypes).
Before training, we remove redundancy by grouping similar sequences into clusters. This is important to reduce bias, as most come from just a few subtypes like H7N9 and H5N1.
Before training, we remove redundancy by grouping similar sequences into clusters. This is important to reduce bias, as most come from just a few subtypes like H7N9 and H5N1.

September 18, 2025 at 1:39 PM
We extracted ~19000 influenza sequences from birds and ~600 zoonotic sequences from humans (only non-seasonal subtypes).
Before training, we remove redundancy by grouping similar sequences into clusters. This is important to reduce bias, as most come from just a few subtypes like H7N9 and H5N1.
Before training, we remove redundancy by grouping similar sequences into clusters. This is important to reduce bias, as most come from just a few subtypes like H7N9 and H5N1.
Lots of ML models can predict human spillover. However for influenza this task is harder because of a) genome segmentation, and b) strong signal within subtype or lineage.
We planned a model training architecture to handle this, ensuring predictions are rooted in virus biology, not shared ancestry.
We planned a model training architecture to handle this, ensuring predictions are rooted in virus biology, not shared ancestry.

September 18, 2025 at 1:39 PM
Lots of ML models can predict human spillover. However for influenza this task is harder because of a) genome segmentation, and b) strong signal within subtype or lineage.
We planned a model training architecture to handle this, ensuring predictions are rooted in virus biology, not shared ancestry.
We planned a model training architecture to handle this, ensuring predictions are rooted in virus biology, not shared ancestry.
That'll be St Elmo - same chap who gives his name to the nautical fire!
July 11, 2025 at 1:54 PM
That'll be St Elmo - same chap who gives his name to the nautical fire!
What is this nightmare of flesh
May 16, 2025 at 1:21 PM
What is this nightmare of flesh
I look forward to reading www.nature.com/articles/s41...!
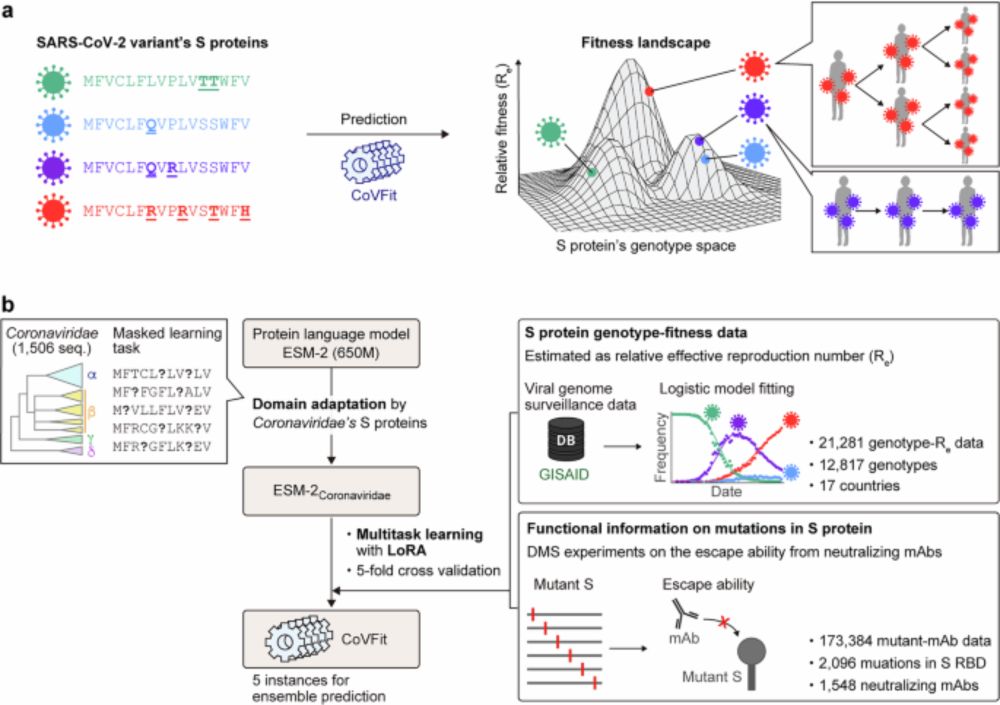
A protein language model for exploring viral fitness landscapes - Nature Communications
Ito et al. present CoVFit, an AI model that predicts variant fitness (transmissibility) from spike protein sequences alone. They further demonstrate its utility in forecasting viral evolution via sing...
www.nature.com
May 15, 2025 at 2:30 PM
I look forward to reading www.nature.com/articles/s41...!
(I know it's the same guy cause the photocopy always caught his shirt cuff in the scan)
May 13, 2025 at 2:03 PM
(I know it's the same guy cause the photocopy always caught his shirt cuff in the scan)

