7/7
7/7
6/7

6/7
5/7

5/7
4/7



4/7
3/7

3/7
2/7

2/7
7/7
7/7
6/7

6/7
5/7

5/7
4/7


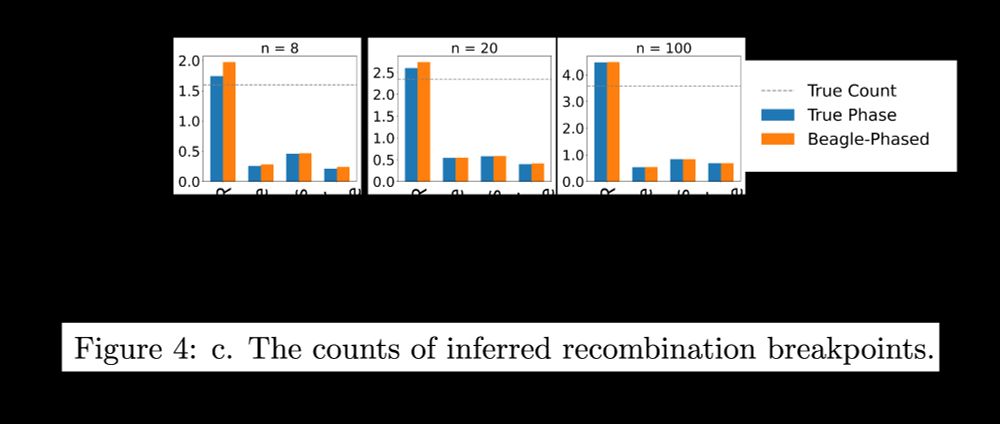
4/7
3/7

3/7
2/7

2/7

