
Teaching machines to learn biology 🧬💻
https://khchao.com/
Mihaela Pertea, @stevensalzberg.bsky.social, Anqi Liu, Alan Mao
🔗 Explore our documentation here: ccb.jhu.edu/openspliceai/ 10/

🔗 Explore our documentation here: ccb.jhu.edu/openspliceai/ 10/

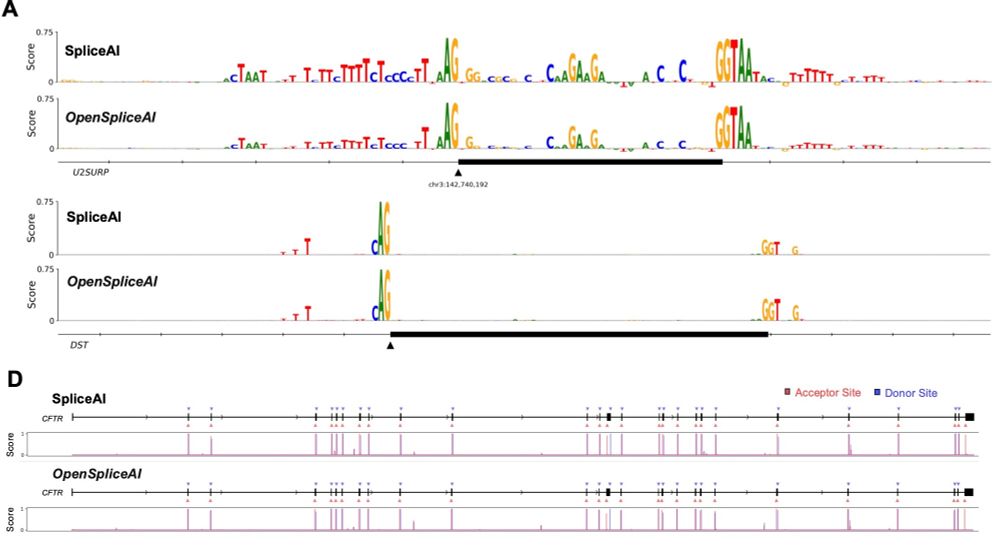




• Create species-specific datasets
• Train custom models
• Calibrate predictions
• Apply transfer learning from human models
• Predict on genes / entire chromosomes
• Assess variant impacts on cryptic splicing
3/

• Create species-specific datasets
• Train custom models
• Calibrate predictions
• Apply transfer learning from human models
• Predict on genes / entire chromosomes
• Assess variant impacts on cryptic splicing
3/
🔗 GitHub: github.com/Kuanhao-Chao...

🔗 GitHub: github.com/Kuanhao-Chao...

