Other soc. media: http://cosoc.com/KSechkar
But even if you couldn't come, you can still watch me embarass myself by trying to write it up as a satirical sketch (link in the next post)

But even if you couldn't come, you can still watch me embarass myself by trying to write it up as a satirical sketch (link in the next post)
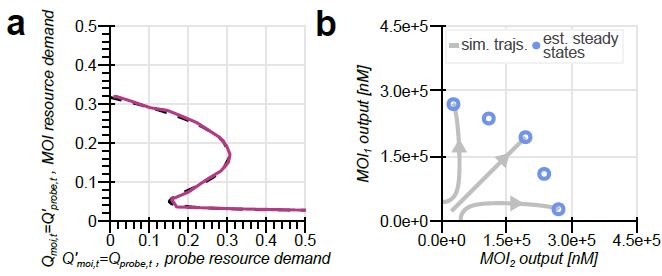
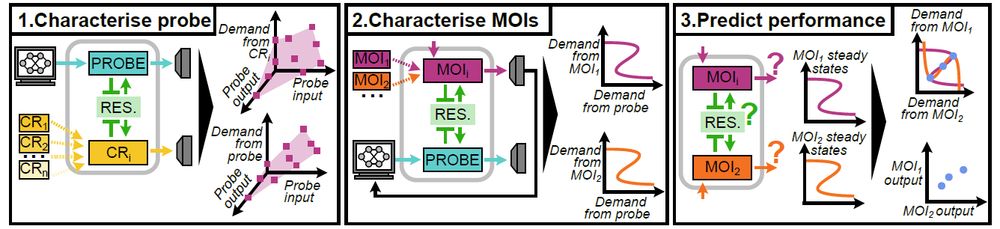
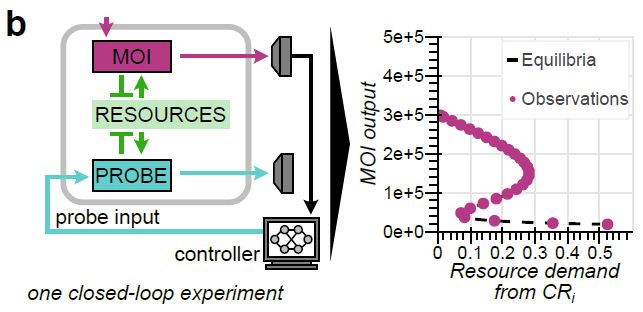

doi.org/10.1101/2025...

doi.org/10.1101/2025...
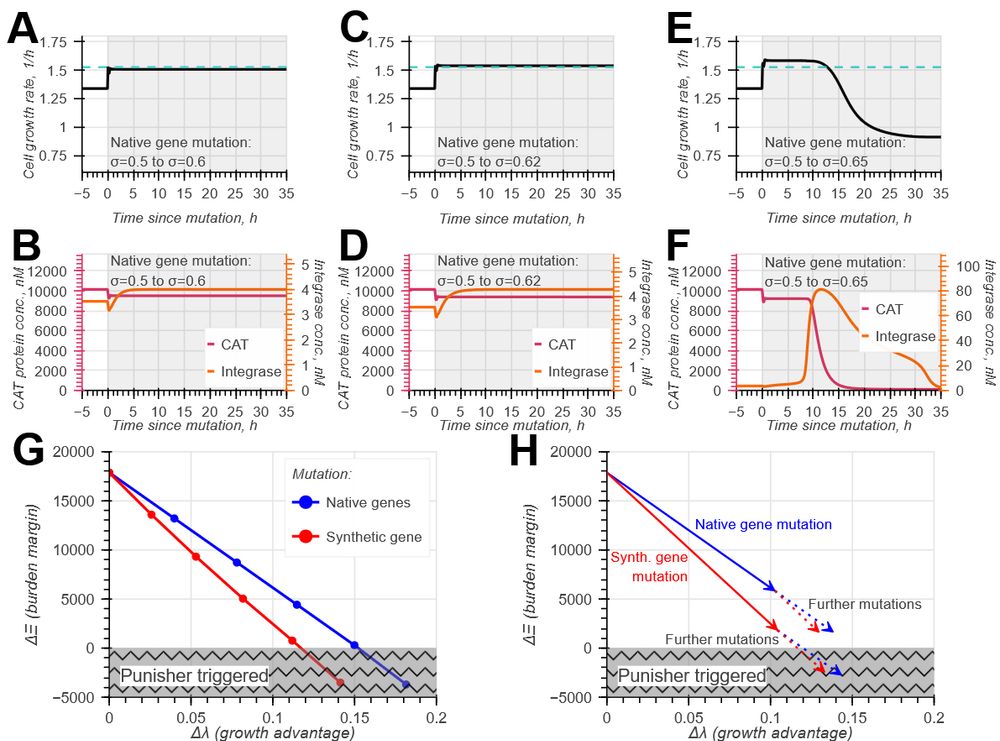
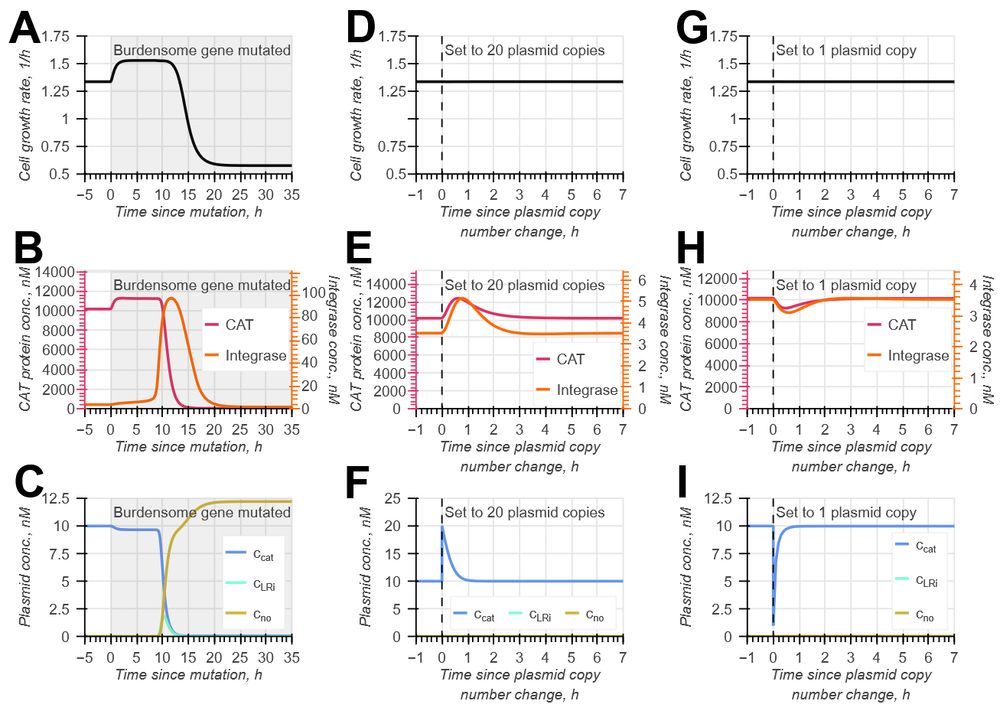
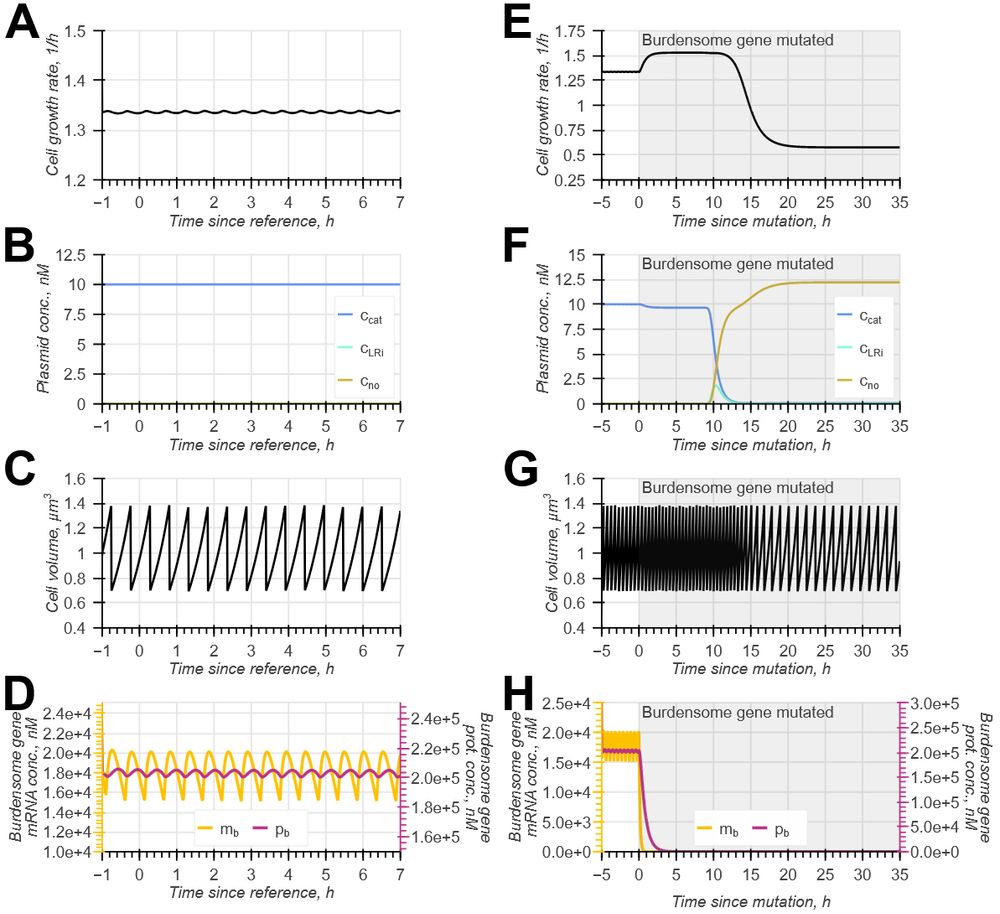
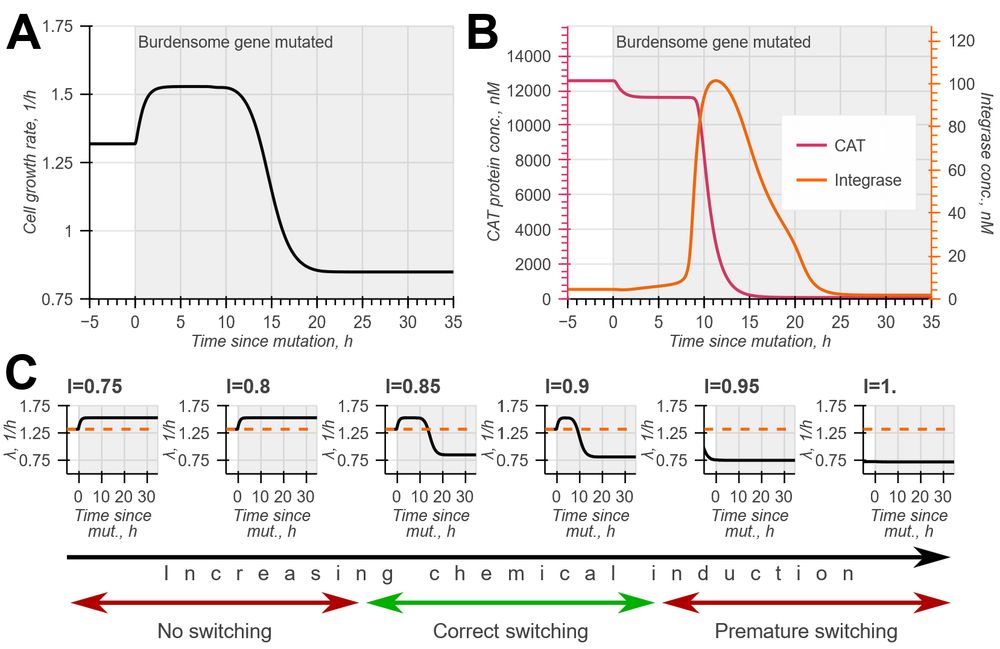
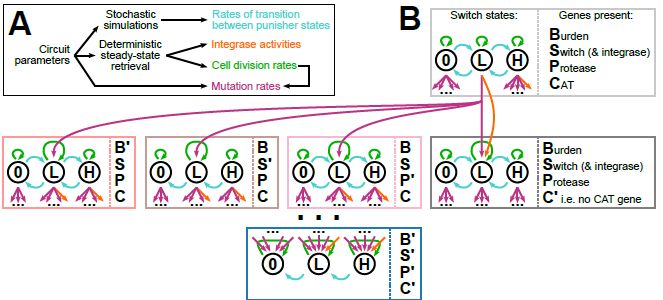
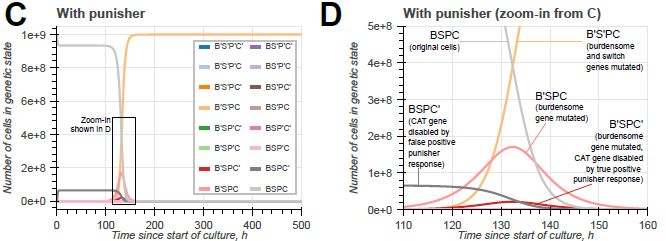
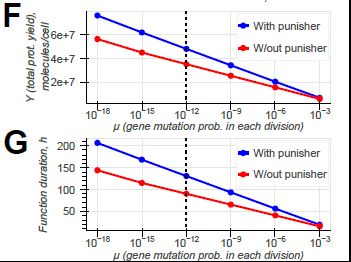
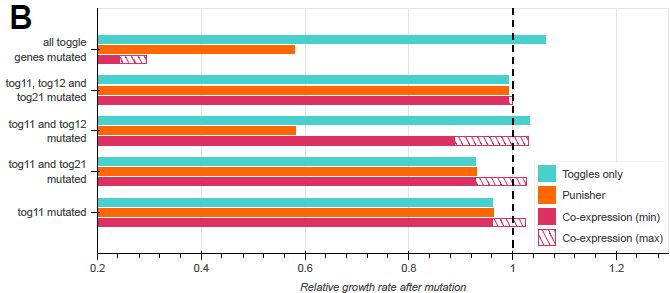
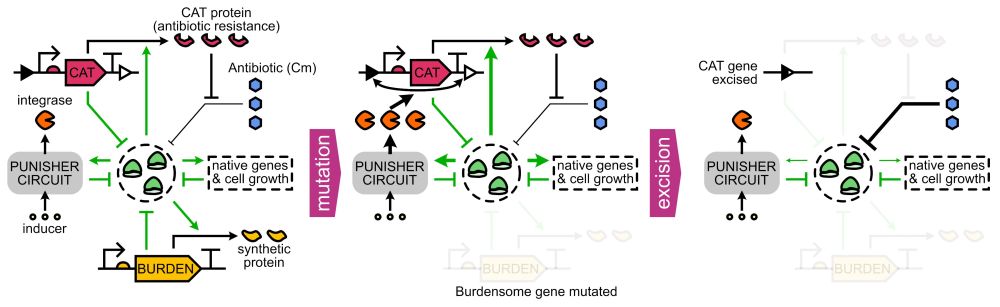
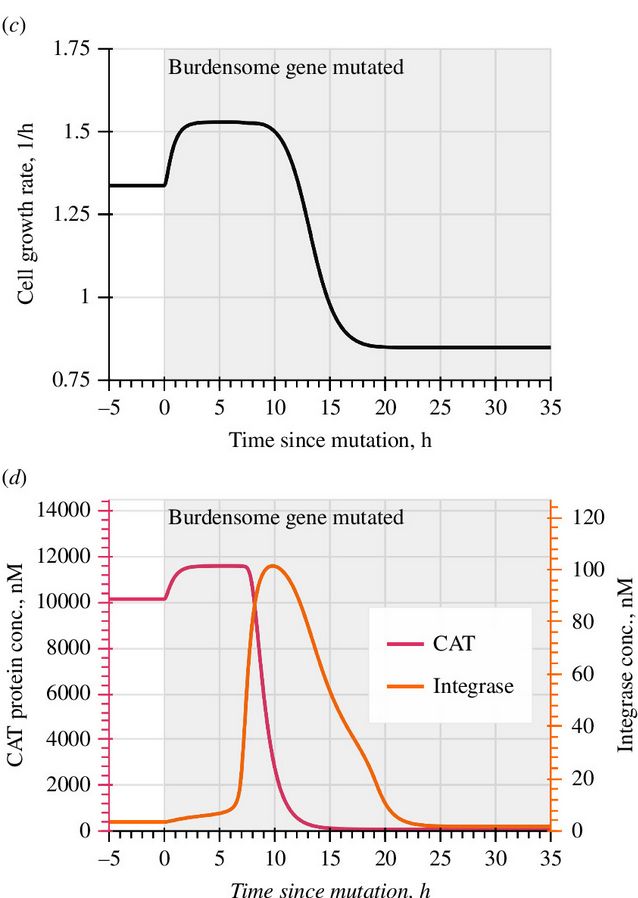
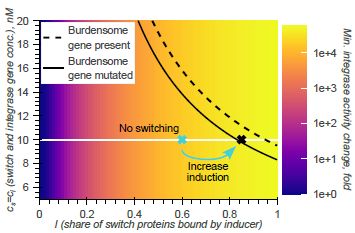
doi.org/10.1098/rsif.2024.0602
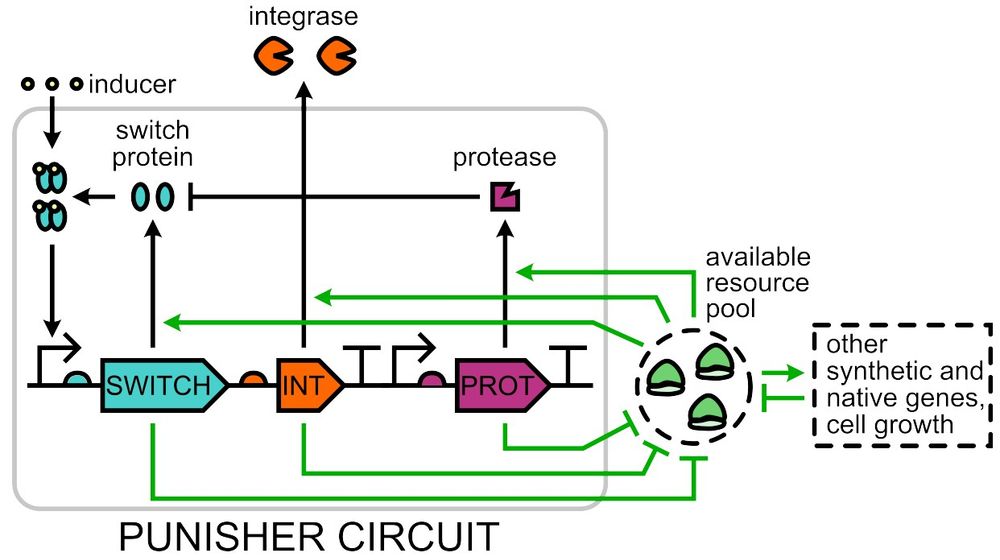
doi.org/10.1098/rsif.2024.0602
Huge thanks to the organisers @engbioirc.bsky.social , to everyone who presented, and to you all for following my live(ish) report!
14/14

Huge thanks to the organisers @engbioirc.bsky.social , to everyone who presented, and to you all for following my live(ish) report!
14/14
4/

4/
2/

2/

Among other things, a very creative way to make artificial cells divide controllably by ligand binding of membranes 2/

Among other things, a very creative way to make artificial cells divide controllably by ligand binding of membranes 2/


