Robson lab - Berlin Institute for Medical Systems Biology (MDC-BIMSB) & Mundlos Lab - Max Planck Institute for Molecular Genetics |
Genome Regulation, Nuclear Envelope, LADs


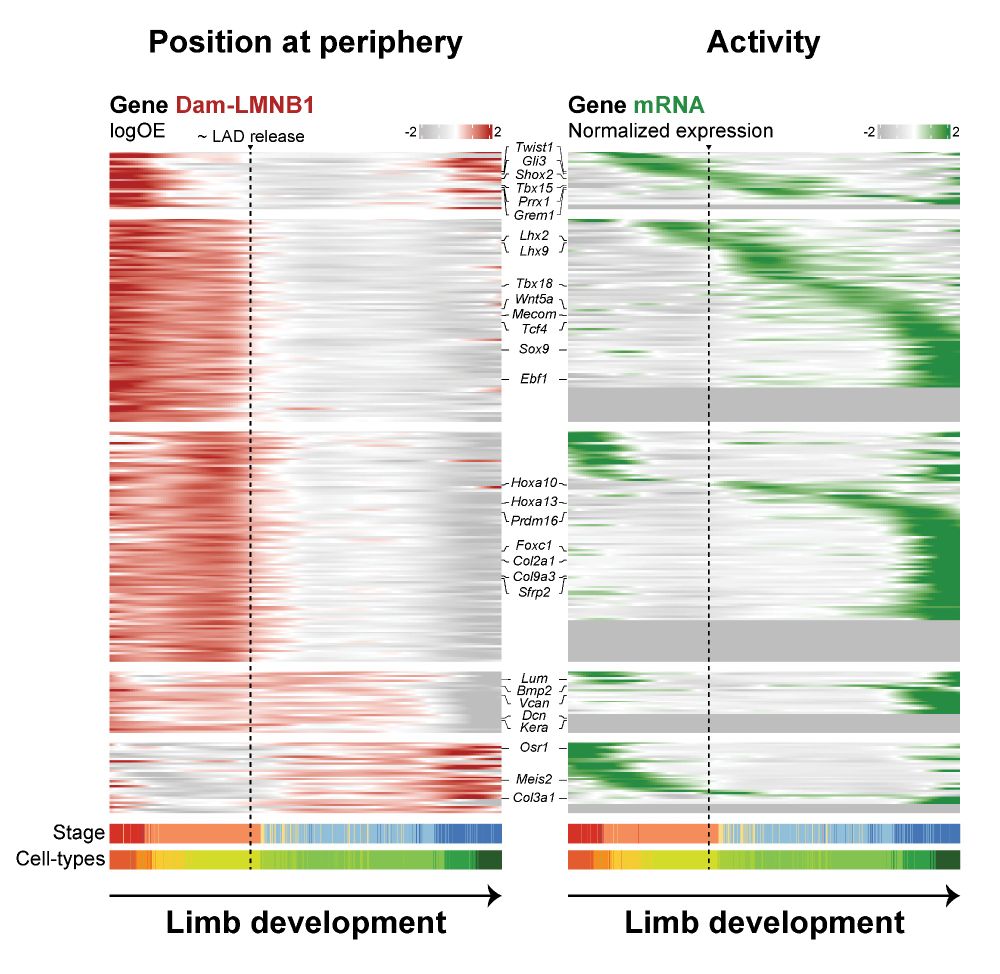

Yuko Sato, Xingchi Yan, Simon Ulrich, @watanyatra.bsky.social, Lothar Schermelleh, Hiroshi Kimura, IrinaSolovei & my supervisor @drmrobson.bsky.social. Thank you for your hard work & support throughout! 🌍🙏
Yuko Sato, Xingchi Yan, Simon Ulrich, @watanyatra.bsky.social, Lothar Schermelleh, Hiroshi Kimura, IrinaSolovei & my supervisor @drmrobson.bsky.social. Thank you for your hard work & support throughout! 🌍🙏
✅ Increase incubation time
✅ Tissue sections (antigen retrieval)
✅ Use lower-affinity antibodies/nanobodies
✅ Check cell-to-cell expression: rims only in high-expression cells = likely artifact

✅ Increase incubation time
✅ Tissue sections (antigen retrieval)
✅ Use lower-affinity antibodies/nanobodies
✅ Check cell-to-cell expression: rims only in high-expression cells = likely artifact

1️⃣ High epitope abundance
2️⃣ High antibody affinity
3️⃣ Low diffusion rate
Any one factor is enough! Potentially affects many antibody–epitope pairs under the right conditions. Examples 👇

1️⃣ High epitope abundance
2️⃣ High antibody affinity
3️⃣ Low diffusion rate
Any one factor is enough! Potentially affects many antibody–epitope pairs under the right conditions. Examples 👇
Y. Sato, X. Yan, S. Urlich, @watanyatra.bsky.social, L. Schermelleh, @gfudenberg.bsky.social, H. Kimura, I. Solovei & my supervisor @mikerobson.bsky.social. Thanks for your hard work & support! 🤝🌍
Y. Sato, X. Yan, S. Urlich, @watanyatra.bsky.social, L. Schermelleh, @gfudenberg.bsky.social, H. Kimura, I. Solovei & my supervisor @mikerobson.bsky.social. Thanks for your hard work & support! 🤝🌍

