
daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org

daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org
www.nature.com/articles/s42...


www.nature.com/articles/s42...



The server: em.kiharalab.org


The server: em.kiharalab.org

Visit the server for easy and accurate modeling at em.kiharalab.org
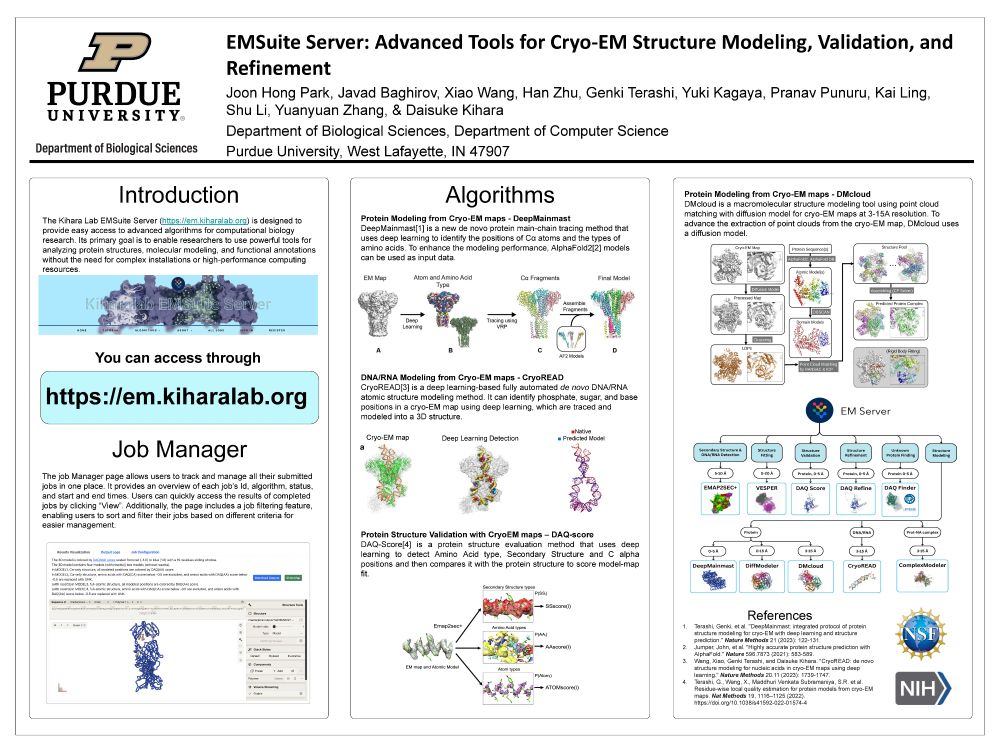
Visit the server for easy and accurate modeling at em.kiharalab.org
Paper: pubs.acs.org/doi/10.1021/...
Github: github.com/kiharalab/Di...
Google Colab: colab.research.google.com/drive/1Q1ecU...
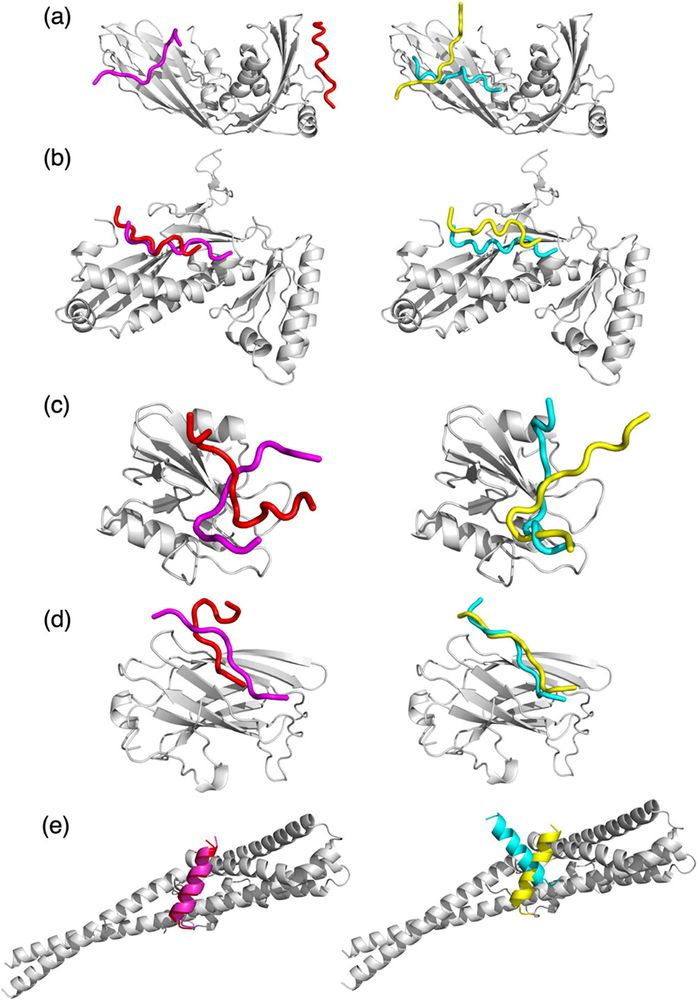
Paper: pubs.acs.org/doi/10.1021/...
Github: github.com/kiharalab/Di...
Google Colab: colab.research.google.com/drive/1Q1ecU...
Each entry shows modeling errors in red on the structure plus chain-wise plots.
Check it out: daqdb.kiharalab.org
You can compute DAQ at: em.kiharalab.org
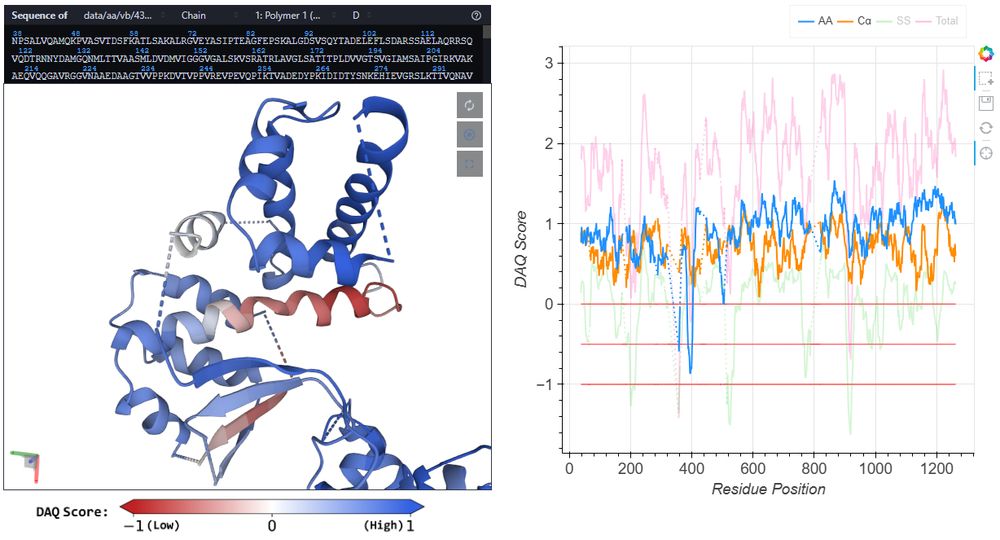
Each entry shows modeling errors in red on the structure plus chain-wise plots.
Check it out: daqdb.kiharalab.org
You can compute DAQ at: em.kiharalab.org
onlinelibrary.wiley.com/doi/10.1002/...
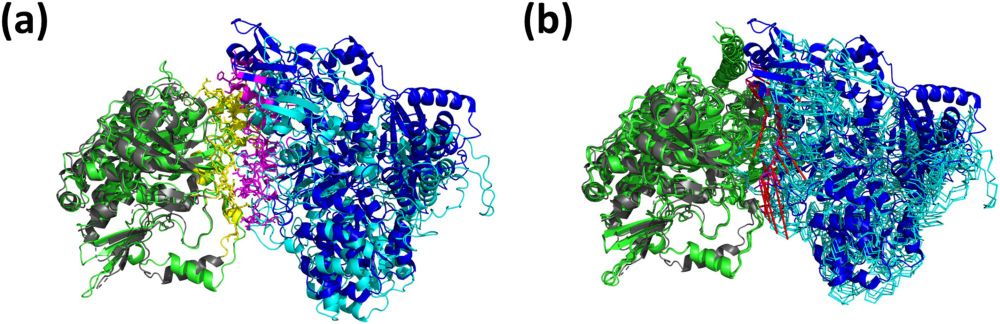
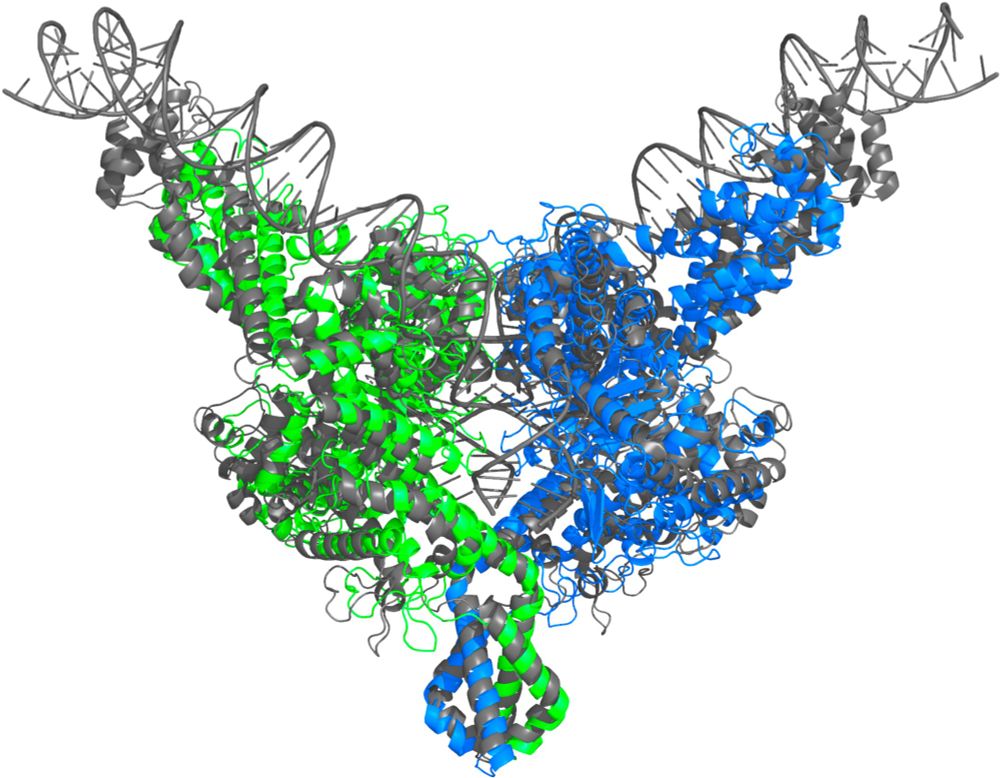
onlinelibrary.wiley.com/doi/10.1002/...

