
If SCTLD involves a symbiont-borne pathogen, this would worsen infection rather than fight it!
Understanding this may be key to detecting and stopping SCTLD earlier.
If SCTLD involves a symbiont-borne pathogen, this would worsen infection rather than fight it!
Understanding this may be key to detecting and stopping SCTLD earlier.
So in HD tissue, Rab5a may reflect:
— symbiont uptake and retention
— early pathogen manipulation
— early host–symbiont dysregulation

So in HD tissue, Rab5a may reflect:
— symbiont uptake and retention
— early pathogen manipulation
— early host–symbiont dysregulation
In Aiptasia, Rab5a localizes to symbiosomes only when they contain viable, newly acquired symbionts (Fig from Chen et al. 2004: doi.org/10.1016/j.bb...)
But there's a twist...

In Aiptasia, Rab5a localizes to symbiosomes only when they contain viable, newly acquired symbionts (Fig from Chen et al. 2004: doi.org/10.1016/j.bb...)
But there's a twist...
One gene stood out: Rab5a, which plays a key role in symbiont uptake and intracellular trafficking in cnidarians.

One gene stood out: Rab5a, which plays a key role in symbiont uptake and intracellular trafficking in cnidarians.
Together, these signal a complete collapse of holobiont homeostasis.


Together, these signal a complete collapse of holobiont homeostasis.




Some of what we found made perfect sense, but some other findings totally surprised us! ‼️
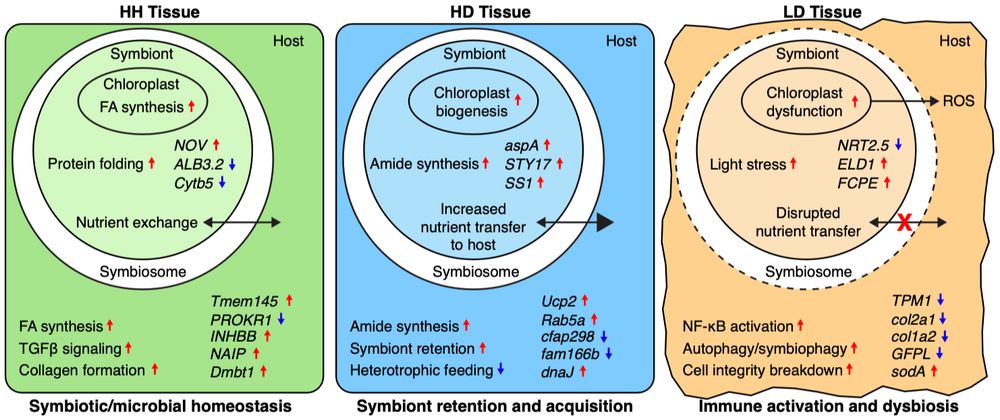
Some of what we found made perfect sense, but some other findings totally surprised us! ‼️
👉 Which genes best distinguish coral health states?
This ML approach ranks genes by their predictive power, and we applied it to both the coral (M. cavernosa) and its main algal symbiont (C. goreaui)

👉 Which genes best distinguish coral health states?
This ML approach ranks genes by their predictive power, and we applied it to both the coral (M. cavernosa) and its main algal symbiont (C. goreaui)
— HH = healthy tissue on a healthy colony
— HD = “healthy” tissue on a diseased colony
— LD = active lesion tissue


— HH = healthy tissue on a healthy colony
— HD = “healthy” tissue on a diseased colony
— LD = active lesion tissue

