Genes and environment profoundly affect the human virome

Genes and environment profoundly affect the human virome







@genomicsdoge.bsky.social @estbiobank.bsky.social @peepkolberg.bsky.social

@genomicsdoge.bsky.social @estbiobank.bsky.social @peepkolberg.bsky.social
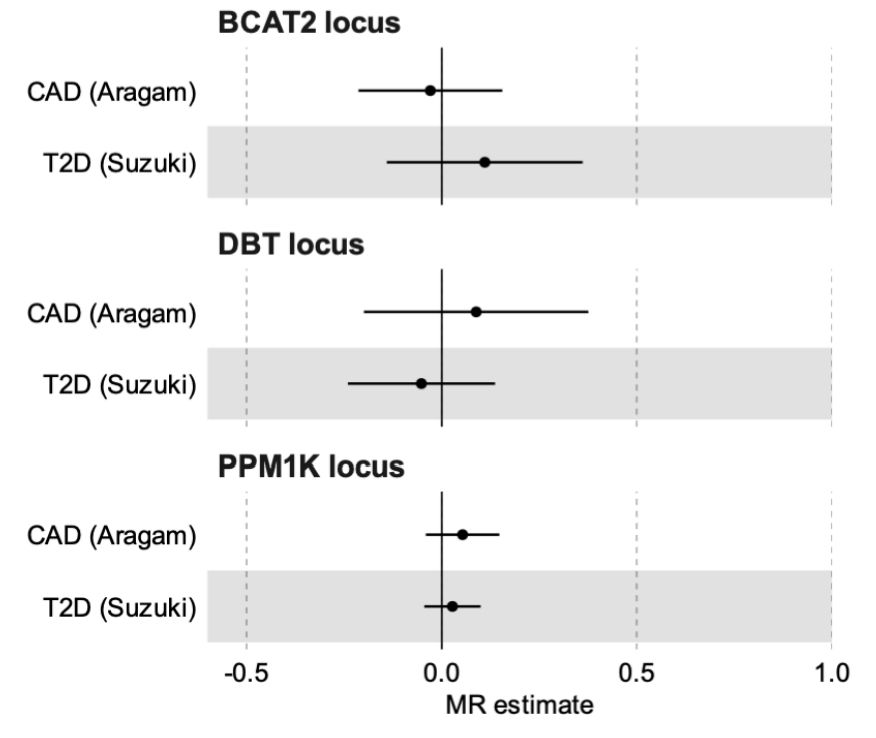
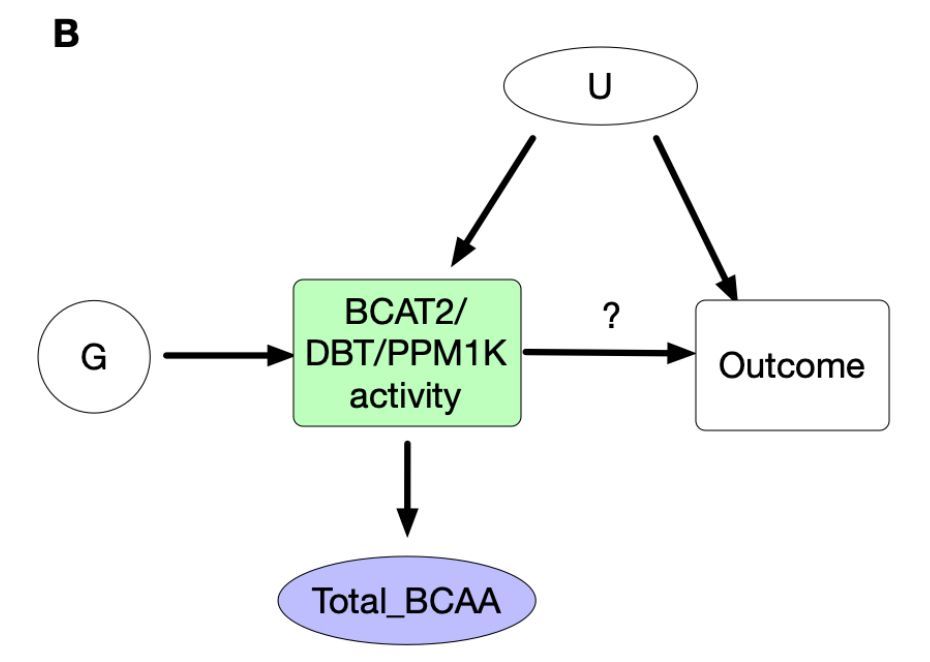
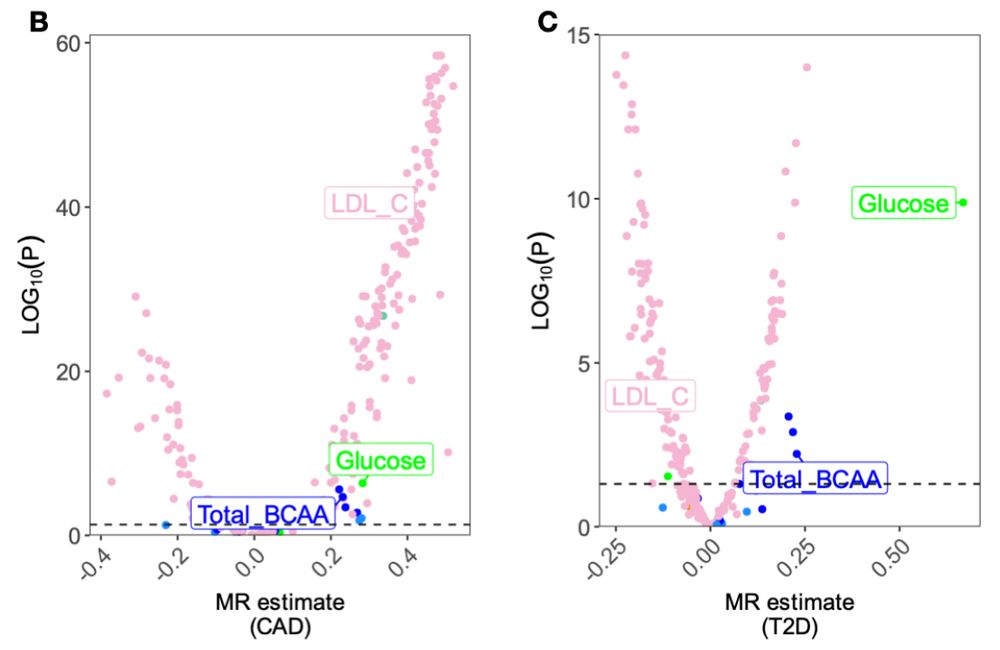

www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...

elixir.ut.ee/eqtl/?rsid=r...

elixir.ut.ee/eqtl/?rsid=r...
spliceailookup.broadinstitute.org#variant=chr1...

spliceailookup.broadinstitute.org#variant=chr1...

What is the causal gene?
What is the mode of action of the causal variant (expression altering, splice altering, missense, etc)?
Hint:

What is the causal gene?
What is the mode of action of the causal variant (expression altering, splice altering, missense, etc)?
Hint:
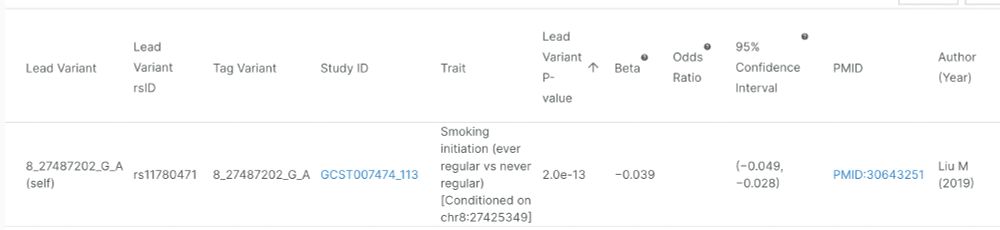

All our top 1000 hits have p < 1e-300. nmrmeta.gi.ut.ee/top_hits

All our top 1000 hits have p < 1e-300. nmrmeta.gi.ut.ee/top_hits




